5'TRU: identification and analysis of translationally regulative 5'untranslated regions in amino acid starved yeast cells
- PMID: 21444828
- PMCID: PMC3108831
- DOI: 10.1074/mcp.M110.003350
5'TRU: identification and analysis of translationally regulative 5'untranslated regions in amino acid starved yeast cells
Abstract
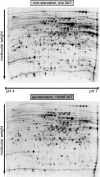
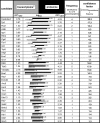
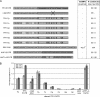
We describe a method to identify and analyze translationally regulative 5'UTRs (5'TRU) in Saccharomyces cerevisiae. Two-dimensional analyses of (35)S-methionine metabolically labeled cells revealed 13 genes and proteins, whose protein biosynthesis is post-transcriptionally up-regulated on amino acid starvation. The 5'UTRs of the respective mRNAs were further investigated. A plasmid-based reporter-testing system was developed to analyze their capability to influence translation dependent on amino acid availability. Most of the 13 candidate 5'UTRs are able to enhance translation independently of amino acids. Two 5'UTRs generally repressed translation, and the 5'UTRs of ENO1, FBA1, and TPI1 specifically up-regulated translation when cells were starved for amino acids. The TPI1-5'UTR exhibited the strongest effect in the testing system, which is consistent with elevated Tpi1p-levels in amino acid starved cells. Bioinformatical analyses support that an unstructured A-rich 5' leader is beneficial for efficient translation when amino acids are scarce. Accordingly, the TPI1-5'UTR was shown to contain an A-rich tract in proximity to the mRNA-initiation codon, required for its amino acid dependent regulatory function.
Figures


Similar articles
-
Sequence and translation initiation properties of the xenopus TGFbeta5, PDGF-A, and PDGF-alpha receptor 5' untranslated regions.Int J Dev Biol. 2000 Dec;44(8):851-9. Int J Dev Biol. 2000. PMID: 11206326
-
Novel splice variants in the 5'UTR of Gtf2i expressed in the rat brain: alternative 5'UTRs and differential expression in the neuronal dendrites.J Neurochem. 2015 Aug;134(3):578-89. doi: 10.1111/jnc.13136. Epub 2015 May 14. J Neurochem. 2015. PMID: 25913238
-
A Novel Method for Gene-Specific Enhancement of Protein Translation by Targeting 5'UTRs of Selected Tumor Suppressors.PLoS One. 2016 May 12;11(5):e0155359. doi: 10.1371/journal.pone.0155359. eCollection 2016. PLoS One. 2016. PMID: 27171412 Free PMC article.
-
A helicase links upstream ORFs and RNA structure.Curr Genet. 2019 Apr;65(2):453-456. doi: 10.1007/s00294-018-0911-z. Epub 2018 Nov 27. Curr Genet. 2019. PMID: 30483885 Free PMC article. Review.
-
Targeting the Iron-Response Elements of the mRNAs for the Alzheimer's Amyloid Precursor Protein and Ferritin to Treat Acute Lead and Manganese Neurotoxicity.Int J Mol Sci. 2019 Feb 25;20(4):994. doi: 10.3390/ijms20040994. Int J Mol Sci. 2019. PMID: 30823541 Free PMC article. Review.
Cited by
-
Translational regulation in response to stress in Saccharomyces cerevisiae.Yeast. 2019 Jan;36(1):5-21. doi: 10.1002/yea.3349. Epub 2018 Sep 3. Yeast. 2019. PMID: 30019452 Free PMC article. Review.
-
Mechanism and Regulation of Protein Synthesis in Saccharomyces cerevisiae.Genetics. 2016 May;203(1):65-107. doi: 10.1534/genetics.115.186221. Genetics. 2016. PMID: 27183566 Free PMC article. Review.
-
RACK1/Asc1p, a ribosomal node in cellular signaling.Mol Cell Proteomics. 2013 Jan;12(1):87-105. doi: 10.1074/mcp.M112.017277. Epub 2012 Oct 15. Mol Cell Proteomics. 2013. PMID: 23071099 Free PMC article.
References
-
- Day D. A., Tuite M. F. (1998) Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J. Endocrinol. 157, 361–371 - PubMed
-
- Holcik M., Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 - PubMed
-
- Hinnebusch A. G. (2000) Mechanism and regulation of initiator methionyl tRNA binding to ribosomes, Translational Control of Gene Expression, pp. 185–243, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
-
- Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

