DNA shape, genetic codes, and evolution
- PMID: 21439813
- PMCID: PMC3112471
- DOI: 10.1016/j.sbi.2011.03.002
DNA shape, genetic codes, and evolution
Abstract
Although the three-letter genetic code that maps nucleotide sequence to protein sequence is well known, there must exist other codes that are embedded in the human genome. Recent work points to sequence-dependent variation in DNA shape as one mechanism by which regulatory and other information could be encoded in DNA. Recent advances include the discovery of shape-dependent recognition of DNA that depends on minor groove width and electrostatics, the existence of overlapping codes in protein-coding regions of the genome, and evolutionary selection for compensatory changes in nucleotide composition that facilitate nucleosome occupancy. It is becoming clear that DNA shape is important to biological function, and therefore will be subject to evolutionary constraint.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
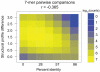
Similar articles
-
The molecular evolution of nucleosome positioning through sequence-dependent deformation of the DNA polymer.J Biomol Struct Dyn. 2010 Jun;27(6):765-80. doi: 10.1080/07391102.2010.10508584. J Biomol Struct Dyn. 2010. PMID: 20232932
-
DNA shape dominates sequence affinity in nucleosome formation.Phys Rev Lett. 2014 Oct 17;113(16):168101. doi: 10.1103/PhysRevLett.113.168101. Epub 2014 Oct 14. Phys Rev Lett. 2014. PMID: 25361282
-
Ubiquitous human 'master' origins of replication are encoded in the DNA sequence via a local enrichment in nucleosome excluding energy barriers.J Phys Condens Matter. 2015 Feb 18;27(6):064102. doi: 10.1088/0953-8984/27/6/064102. Epub 2015 Jan 7. J Phys Condens Matter. 2015. PMID: 25563930
-
Thirty years of multiple sequence codes.Genomics Proteomics Bioinformatics. 2011 Apr;9(1-2):1-6. doi: 10.1016/S1672-0229(11)60001-6. Genomics Proteomics Bioinformatics. 2011. PMID: 21641556 Free PMC article. Review.
-
Cracking the chromatin code: precise rule of nucleosome positioning.Phys Life Rev. 2011 Mar;8(1):39-50. doi: 10.1016/j.plrev.2011.01.004. Epub 2011 Jan 19. Phys Life Rev. 2011. PMID: 21295529 Review.
Cited by
-
Computational models for large-scale simulations of facilitated diffusion.Mol Biosyst. 2012 Nov;8(11):2815-27. doi: 10.1039/c2mb25201e. Epub 2012 Aug 15. Mol Biosyst. 2012. PMID: 22892851 Free PMC article. Review.
-
Thermodynamics of nucleic acid "shape readout" by an aminosugar.Biochemistry. 2011 Oct 25;50(42):9088-113. doi: 10.1021/bi201077h. Epub 2011 Oct 3. Biochemistry. 2011. PMID: 21863895 Free PMC article.
-
TFBSshape: a motif database for DNA shape features of transcription factor binding sites.Nucleic Acids Res. 2014 Jan;42(Database issue):D148-55. doi: 10.1093/nar/gkt1087. Epub 2013 Nov 7. Nucleic Acids Res. 2014. PMID: 24214955 Free PMC article.
-
Absence of a simple code: how transcription factors read the genome.Trends Biochem Sci. 2014 Sep;39(9):381-99. doi: 10.1016/j.tibs.2014.07.002. Epub 2014 Aug 14. Trends Biochem Sci. 2014. PMID: 25129887 Free PMC article. Review.
-
Recruitment of CRISPR-Cas systems by Tn7-like transposons.Proc Natl Acad Sci U S A. 2017 Aug 29;114(35):E7358-E7366. doi: 10.1073/pnas.1709035114. Epub 2017 Aug 15. Proc Natl Acad Sci U S A. 2017. PMID: 28811374 Free PMC article.
References
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Dobzhansky T. Nothing in biology makes sense except in the light of evolution. American Biol Teacher. 1973;35:125–129.
-
-
Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. ••This paper introduced a new method for assessing evolutionary constraint based on DNA shape, and showed that more than 10% of the human genome is under selection for structure. Of particular significance is the finding that a high proportion of functional genomic elements occur in regions that are under structural constraint.
-
-
-
Itzkovitz S, Hodis E, Segal E. Overlapping codes within protein-coding sequences. Genome Res. 2010;20:1582–1589. •• This study presents a clever method to measure information content in coding sequences. The authors found that across diverse phyla coding sequences encode information beyond the standard genetic code.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

