Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression
- PMID: 21436584
- PMCID: PMC3069786
- DOI: 10.1172/JCI45284
Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression
Abstract
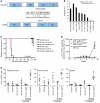
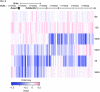
Acute promyelocytic leukemia (APL) is a subtype of acute myeloid leukemia (AML). It is characterized by the t(15;17)(q22;q11.2) chromosomal translocation that creates the promyelocytic leukemia-retinoic acid receptor α (PML-RARA) fusion oncogene. Although this fusion oncogene is known to initiate APL in mice, other cooperating mutations, as yet ill defined, are important for disease pathogenesis. To identify these, we used a mouse model of APL, whereby PML-RARA expressed in myeloid cells leads to a myeloproliferative disease that ultimately evolves into APL. Sequencing of a mouse APL genome revealed 3 somatic, nonsynonymous mutations relevant to APL pathogenesis, of which 1 (Jak1 V657F) was found to be recurrent in other affected mice. This mutation was identical to the JAK1 V658F mutation previously found in human APL and acute lymphoblastic leukemia samples. Further analysis showed that JAK1 V658F cooperated in vivo with PML-RARA, causing a rapidly fatal leukemia in mice. We also discovered a somatic 150-kb deletion involving the lysine (K)-specific demethylase 6A (Kdm6a, also known as Utx) gene, in the mouse APL genome. Similar deletions were observed in 3 out of 14 additional mouse APL samples and 1 out of 150 human AML samples. In conclusion, whole genome sequencing of mouse cancer genomes can provide an unbiased and comprehensive approach for discovering functionally relevant mutations that are also present in human leukemias.
Figures




Comment in
-
Finding a needle in a haystack: whole genome sequencing and mutation discovery in murine models.J Clin Invest. 2011 Apr;121(4):1255-8. doi: 10.1172/JCI57200. Epub 2011 Mar 23. J Clin Invest. 2011. PMID: 21436577 Free PMC article.
Similar articles
-
Finding a needle in a haystack: whole genome sequencing and mutation discovery in murine models.J Clin Invest. 2011 Apr;121(4):1255-8. doi: 10.1172/JCI57200. Epub 2011 Mar 23. J Clin Invest. 2011. PMID: 21436577 Free PMC article.
-
Recurrent RARB Translocations in Acute Promyelocytic Leukemia Lacking RARA Translocation.Cancer Res. 2018 Aug 15;78(16):4452-4458. doi: 10.1158/0008-5472.CAN-18-0840. Epub 2018 Jun 19. Cancer Res. 2018. PMID: 29921692
-
Missense mutations in the PML/RARalpha ligand binding domain in ATRA-resistant As(2)O(3) sensitive relapsed acute promyelocytic leukemia.Haematologica. 1999 Nov;84(11):963-8. Haematologica. 1999. PMID: 10553155
-
Therapy-induced PML/RARA proteolysis and acute promyelocytic leukemia cure.Clin Cancer Res. 2009 Oct 15;15(20):6321-6. doi: 10.1158/1078-0432.CCR-09-0209. Epub 2009 Oct 6. Clin Cancer Res. 2009. PMID: 19808868 Review.
-
Molecular analysis of the t(15;17) translocation in acute promyelocytic leukaemia.Baillieres Clin Haematol. 1992 Oct;5(4):833-56. doi: 10.1016/s0950-3536(11)80048-x. Baillieres Clin Haematol. 1992. PMID: 1339190 Review.
Cited by
-
The genetics of splicing in neuroblastoma.Cancer Discov. 2015 Apr;5(4):380-95. doi: 10.1158/2159-8290.CD-14-0892. Epub 2015 Jan 30. Cancer Discov. 2015. PMID: 25637275 Free PMC article.
-
A comprehensive review of genetic alterations and molecular targeted therapies for the implementation of personalized medicine in acute myeloid leukemia.J Mol Med (Berl). 2020 Aug;98(8):1069-1091. doi: 10.1007/s00109-020-01944-5. Epub 2020 Jul 3. J Mol Med (Berl). 2020. PMID: 32620999 Review.
-
The Cross Marks the Spot: The Emerging Role of JmjC Domain-Containing Proteins in Myeloid Malignancies.Biomolecules. 2021 Dec 20;11(12):1911. doi: 10.3390/biom11121911. Biomolecules. 2021. PMID: 34944554 Free PMC article. Review.
-
Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation.Blood. 2015 Jan 22;125(4):619-28. doi: 10.1182/blood-2014-08-594564. Blood. 2015. PMID: 25416276 Free PMC article.
-
How animal models of leukaemias have already benefited patients.Mol Oncol. 2013 Apr;7(2):224-31. doi: 10.1016/j.molonc.2013.01.006. Epub 2013 Feb 11. Mol Oncol. 2013. PMID: 23453906 Free PMC article. Review.
References
-
- Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML–RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89(2):376–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

