βTrCP regulates BMI1 protein turnover via ubiquitination and degradation
- PMID: 21430439
- PMCID: PMC3117138
- DOI: 10.4161/cc.10.8.15372
βTrCP regulates BMI1 protein turnover via ubiquitination and degradation
Abstract
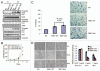
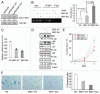
The polycomb group protein BMI1 has been linked to proliferation, senescence, cancer progression and stem cell phenotype. At present, very little is known about its regulation. Here, we report that BMI1 contains a functional recognition motif for the F box protein βTrCP, which regulates ubiquitination and proteasome-mediated degradation of various proteins. We show that overexpression of wild-type βTrCP but not the ΔF mutant of it promotes BMI1 ubiquitination and degradation, and knockdown of βTrCP results in increased expression of BMI1. Furthermore, a mutant of BMI1 with an altered βTrCP recognition motif is much more stable than wild-type BMI1. We also show that wild-type BMI1 but not the mutant BMI1 interacts with βTrCP. Accordingly, compared to wild-type BMI1, mutant protein exhibited increased pro-oncogenic activity. In summary, our findings suggest that βTrCP regulates turnover of BMI1 and its function relevant to oncogenesis, cellular senescence and aging.
Figures



Comment in
-
BMI1: a target of βTrCP.Cell Cycle. 2011 Jun 15;10(12):1893-4. doi: 10.4161/cc.10.12.15691. Epub 2011 Jun 15. Cell Cycle. 2011. PMID: 21577051 No abstract available.
-
BMI1 suffers a degrading experience.Cell Cycle. 2011 Jun 15;10(12):1894-5. doi: 10.4161/cc.10.12.15688. Epub 2011 Jun 15. Cell Cycle. 2011. PMID: 21593587 Free PMC article. No abstract available.
Similar articles
-
The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex - a potentially new drug target.PLoS One. 2009 Dec 1;4(12):e8104. doi: 10.1371/journal.pone.0008104. PLoS One. 2009. PMID: 19956605 Free PMC article.
-
BMI1: a target of βTrCP.Cell Cycle. 2011 Jun 15;10(12):1893-4. doi: 10.4161/cc.10.12.15691. Epub 2011 Jun 15. Cell Cycle. 2011. PMID: 21577051 No abstract available.
-
The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors.Cell Cycle. 2010 Jul 1;9(13):2663-73. doi: 10.4161/cc.9.13.12147. Cell Cycle. 2010. PMID: 20543557 Free PMC article.
-
BMI1 as a novel target for drug discovery in cancer.J Cell Biochem. 2011 Oct;112(10):2729-41. doi: 10.1002/jcb.23234. J Cell Biochem. 2011. PMID: 21678481 Review.
-
Bmi1, stem cells, and senescence regulation.J Clin Invest. 2004 Jan;113(2):175-9. doi: 10.1172/JCI20800. J Clin Invest. 2004. PMID: 14722607 Free PMC article. Review.
Cited by
-
Targeting cancer stemness mediated by BMI1 and MCL1 for non-small cell lung cancer treatment.J Cell Mol Med. 2022 Aug;26(15):4305-4321. doi: 10.1111/jcmm.17453. Epub 2022 Jul 6. J Cell Mol Med. 2022. PMID: 35794816 Free PMC article.
-
Acute myeloid leukemia driven by the CALM-AF10 fusion gene is dependent on BMI1.Exp Hematol. 2019 Jun;74:42-51.e3. doi: 10.1016/j.exphem.2019.04.003. Epub 2019 May 13. Exp Hematol. 2019. PMID: 31022428 Free PMC article.
-
Fbxo45 inhibits calcium-sensitive proteolysis of N-cadherin and promotes neuronal differentiation.J Biol Chem. 2014 Oct 10;289(41):28448-59. doi: 10.1074/jbc.M114.561241. Epub 2014 Aug 20. J Biol Chem. 2014. PMID: 25143387 Free PMC article.
-
The role of aberrant proteolysis in lymphomagenesis.Curr Opin Hematol. 2015 Jul;22(4):369-78. doi: 10.1097/MOH.0000000000000156. Curr Opin Hematol. 2015. PMID: 26049759 Free PMC article. Review.
-
BMI1 suffers a degrading experience.Cell Cycle. 2011 Jun 15;10(12):1894-5. doi: 10.4161/cc.10.12.15688. Epub 2011 Jun 15. Cell Cycle. 2011. PMID: 21593587 Free PMC article. No abstract available.
References
-
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. - PubMed
-
- Kim JH, Yoon SY, Jeong SH, Kim SY, Moon SK, Joo JH, et al. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast. 2004;13:383–388. - PubMed
-
- Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
