Improving RNA-Seq expression estimates by correcting for fragment bias
- PMID: 21410973
- PMCID: PMC3129672
- DOI: 10.1186/gb-2011-12-3-r22
Improving RNA-Seq expression estimates by correcting for fragment bias
Abstract
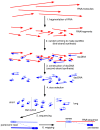
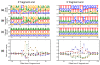
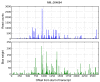
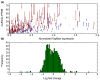
The biochemistry of RNA-Seq library preparation results in cDNA fragments that are not uniformly distributed within the transcripts they represent. This non-uniformity must be accounted for when estimating expression levels, and we show how to perform the needed corrections using a likelihood based approach. We find improvements in expression estimates as measured by correlation with independently performed qRT-PCR and show that correction of bias leads to improved replicability of results across libraries and sequencing technologies.
Figures
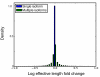
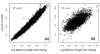
Similar articles
-
Peregrine: A rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material.RNA Biol. 2013 Apr;10(4):502-15. doi: 10.4161/rna.24284. Epub 2013 Apr 1. RNA Biol. 2013. PMID: 23558773 Free PMC article.
-
Library preparation methods for next-generation sequencing: tone down the bias.Exp Cell Res. 2014 Mar 10;322(1):12-20. doi: 10.1016/j.yexcr.2014.01.008. Epub 2014 Jan 15. Exp Cell Res. 2014. PMID: 24440557 Review.
-
Optimization for Sequencing and Analysis of Degraded FFPE-RNA Samples.J Vis Exp. 2020 Jun 8;(160):10.3791/61060. doi: 10.3791/61060. J Vis Exp. 2020. PMID: 32568231 Free PMC article.
-
Mixture models reveal multiple positional bias types in RNA-Seq data and lead to accurate transcript concentration estimates.PLoS Comput Biol. 2017 May 15;13(5):e1005515. doi: 10.1371/journal.pcbi.1005515. eCollection 2017 May. PLoS Comput Biol. 2017. PMID: 28505151 Free PMC article.
-
Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing.Curr Protoc Mol Biol. 2014 Jul 1;107:4.22.1-4.22.17. doi: 10.1002/0471142727.mb0422s107. Curr Protoc Mol Biol. 2014. PMID: 24984854 Free PMC article. Review.
Cited by
-
Anti-bias training for (sc)RNA-seq: experimental and computational approaches to improve precision.Brief Bioinform. 2021 Nov 5;22(6):bbab148. doi: 10.1093/bib/bbab148. Brief Bioinform. 2021. PMID: 33959753 Free PMC article.
-
Whole-genome mapping of 5' RNA ends in bacteria by tagged sequencing: a comprehensive view in Enterococcus faecalis.RNA. 2015 May;21(5):1018-30. doi: 10.1261/rna.048470.114. Epub 2015 Mar 3. RNA. 2015. PMID: 25737579 Free PMC article.
-
DBATE: database of alternative transcripts expression.Database (Oxford). 2013 Jul 9;2013:bat050. doi: 10.1093/database/bat050. Print 2013. Database (Oxford). 2013. PMID: 23842462 Free PMC article.
-
Nervonic acid improves fat transplantation by promoting adipogenesis and angiogenesis.Int J Mol Med. 2024 Dec;54(6):108. doi: 10.3892/ijmm.2024.5432. Epub 2024 Oct 4. Int J Mol Med. 2024. PMID: 39364738 Free PMC article.
-
Detecting differential usage of exons from RNA-seq data.Genome Res. 2012 Oct;22(10):2008-17. doi: 10.1101/gr.133744.111. Epub 2012 Jun 21. Genome Res. 2012. PMID: 22722343 Free PMC article.
References
-
- Paşaniuc B, Zaitlen N, Halperin E. In: Research in Computational Molecular Biology. Berger B, editor. Berlin/Heidelberg: Springer; 2010. Accurate estimation of expression levels of homologous genes in RNA-seq experiments. pp. 397–409. [Lecture Notes in Computer Science, vol 6044.] - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

