Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening
- PMID: 21398570
- PMCID: PMC3082273
- DOI: 10.1105/tpc.110.081273
Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening
Abstract
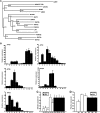
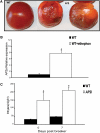
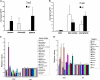
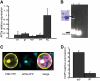
Fruit ripening in tomato (Solanum lycopersicum) requires the coordination of both developmental cues as well as the plant hormone ethylene. Although the role of ethylene in mediating climacteric ripening has been established, knowledge regarding the developmental regulators that modulate the involvement of ethylene in tomato fruit ripening is still lacking. Here, we show that the tomato APETALA2a (AP2a) transcription factor regulates fruit ripening via regulation of ethylene biosynthesis and signaling. RNA interference (RNAi)-mediated repression of AP2a resulted in alterations in fruit shape, orange ripe fruits, and altered carotenoid accumulation. Microarray expression analyses of the ripe AP2 RNAi fruits showed altered expression of genes involved in various metabolic pathways, such as the phenylpropanoid and carotenoid pathways, as well as in hormone synthesis and perception. Genes involved in chromoplast differentiation and other ripening-associated processes were also differentially expressed, but softening and ethylene biosynthesis occurred in the transgenic plants. Ripening regulators RIPENING-INHIBITOR, NON-RIPENING, and COLORLESS NON-RIPENING (CNR) function upstream of AP2a and positively regulate its expression. In the pericarp of AP2 RNAi fruits, mRNA levels of CNR were elevated, indicating that AP2a and CNR are part of a negative feedback loop in the regulation of ripening. Moreover, we demonstrated that CNR binds to the promoter of AP2a in vitro.
Figures

Similar articles
-
A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation.Plant Cell Physiol. 2014 Jan;55(1):119-35. doi: 10.1093/pcp/pct162. Epub 2013 Nov 20. Plant Cell Physiol. 2014. PMID: 24265273
-
A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening.Plant J. 2010 Dec;64(6):936-47. doi: 10.1111/j.1365-313X.2010.04384.x. Epub 2010 Nov 17. Plant J. 2010. PMID: 21143675
-
TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network.Plant J. 2009 Dec;60(6):1081-95. doi: 10.1111/j.1365-313X.2009.04064.x. Plant J. 2009. PMID: 19891701
-
Transcriptional control of fleshy fruit development and ripening.J Exp Bot. 2014 Aug;65(16):4527-41. doi: 10.1093/jxb/eru316. J Exp Bot. 2014. PMID: 25080453 Review.
-
Regulation of carotenoid formation during tomato fruit ripening and development.J Exp Bot. 2002 Oct;53(377):2107-13. doi: 10.1093/jxb/erf059. J Exp Bot. 2002. PMID: 12324534 Review.
Cited by
-
Reactive oxygen species involved in regulating fruit senescence and fungal pathogenicity.Plant Mol Biol. 2013 Aug;82(6):593-602. doi: 10.1007/s11103-013-0035-2. Epub 2013 Mar 21. Plant Mol Biol. 2013. PMID: 23515879 Review.
-
Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato.Plant Physiol. 2012 Jul;159(3):1086-98. doi: 10.1104/pp.112.197483. Epub 2012 May 22. Plant Physiol. 2012. PMID: 22623517 Free PMC article.
-
Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172.Plant Mol Biol. 2012 Mar;78(4-5):417-29. doi: 10.1007/s11103-012-9877-2. Plant Mol Biol. 2012. PMID: 22290408
-
Characterization of a Potential Ripening Regulator, SlNAC3, from Solanum Lycopersicum.Open Life Sci. 2018 Dec 31;13:518-526. doi: 10.1515/biol-2018-0062. eCollection 2018 Jan. Open Life Sci. 2018. PMID: 33817122 Free PMC article.
-
TM3 and STM3 Promote Flowering Together with FUL2 and MBP20, but Act Antagonistically in Inflorescence Branching in Tomato.Plants (Basel). 2023 Jul 25;12(15):2754. doi: 10.3390/plants12152754. Plants (Basel). 2023. PMID: 37570908 Free PMC article.
References
-
- Abuqamar S., Luo H., Laluk K., Mickelbart M.V., Mengiste T. (2009). Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 58: 347–360 - PubMed
-
- Aker J., Borst J.W., Karlova R., de Vries S. (2006). The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J. Struct. Biol. 156: 62–71 - PubMed
-
- Alexander L., Grierson D. (2002). Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 53: 2039–2055 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

