ESCRT-III protein requirements for HIV-1 budding
- PMID: 21396898
- PMCID: PMC3070458
- DOI: 10.1016/j.chom.2011.02.004
ESCRT-III protein requirements for HIV-1 budding
Abstract
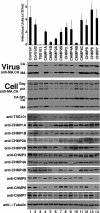
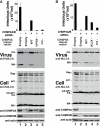
Two early-acting components of the cellular ESCRT pathway, ESCRT-I and ALIX, participate directly in HIV-1 budding. The membrane fission activities of ESCRT-III subunits are also presumably required, but humans express 11 different CHMP/ESCRT-III proteins whose functional contributions are not yet clear. We therefore depleted cells of each of the different CHMP proteins and protein families and examined the effects on HIV-1 budding. Virus release was profoundly inhibited by codepletion of either CHMP2 or CHMP4 family members, resulting in ≥100-fold titer reductions. CHMP2A and CHMP4B proteins bound one another, and this interaction was required for budding. By contrast, virus release was reduced only modestly by depletion of CHMP3 and CHMP1 proteins (2- to 8-fold titer reductions) and was unaffected by depletion of other human ESCRT-III proteins. HIV-1 budding therefore requires only a subset of the known human ESCRT-III proteins, with the CHMP2 and CHMP4 families playing key functional roles.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Essential ingredients for HIV-1 budding.Cell Host Microbe. 2011 Mar 17;9(3):172-174. doi: 10.1016/j.chom.2011.03.005. Cell Host Microbe. 2011. PMID: 21402355
Similar articles
-
In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters.Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16928-33. doi: 10.1073/pnas.1211759109. Epub 2012 Oct 1. Proc Natl Acad Sci U S A. 2012. PMID: 23027949 Free PMC article.
-
Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites.PLoS Pathog. 2015 Feb 24;11(2):e1004677. doi: 10.1371/journal.ppat.1004677. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25710462 Free PMC article.
-
Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A.J Virol. 2012 Apr;86(7):3746-56. doi: 10.1128/JVI.06539-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258254 Free PMC article.
-
The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review.
-
The role of cellular factors in promoting HIV budding.J Mol Biol. 2011 Jul 22;410(4):525-33. doi: 10.1016/j.jmb.2011.04.055. J Mol Biol. 2011. PMID: 21762798 Free PMC article. Review.
Cited by
-
Alix-mediated assembly of the actomyosin-tight junction polarity complex preserves epithelial polarity and epithelial barrier.Nat Commun. 2016 Jun 23;7:11876. doi: 10.1038/ncomms11876. Nat Commun. 2016. PMID: 27336173 Free PMC article.
-
How to get out: ssRNA enveloped viruses and membrane fission.Curr Opin Virol. 2013 Apr;3(2):159-67. doi: 10.1016/j.coviro.2013.03.011. Epub 2013 Apr 11. Curr Opin Virol. 2013. PMID: 23583788 Free PMC article. Review.
-
A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):E8900-E8908. doi: 10.1073/pnas.1805504115. Epub 2018 Sep 4. Proc Natl Acad Sci U S A. 2018. PMID: 30181294 Free PMC article.
-
ESCRT requirements for EIAV budding.Retrovirology. 2013 Oct 9;10:104. doi: 10.1186/1742-4690-10-104. Retrovirology. 2013. PMID: 24107264 Free PMC article.
-
Vaccinia virus hijacks ESCRT-mediated multivesicular body formation for virus egress.Life Sci Alliance. 2021 Jun 18;4(8):e202000910. doi: 10.26508/lsa.202000910. Print 2021 Aug. Life Sci Alliance. 2021. PMID: 34145027 Free PMC article.
References
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Babst M, Katzmann D, Estepa-Sabal E, Meerloo T, Emr S. Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

