Ordered and dynamic assembly of single spliceosomes
- PMID: 21393538
- PMCID: PMC3086749
- DOI: 10.1126/science.1198830
Ordered and dynamic assembly of single spliceosomes
Abstract
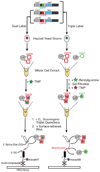
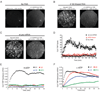
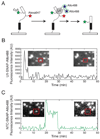
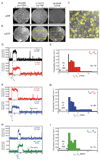
The spliceosome is the complex macromolecular machine responsible for removing introns from precursors to messenger RNAs (pre-mRNAs). We combined yeast genetic engineering, chemical biology, and multiwavelength fluorescence microscopy to follow assembly of single spliceosomes in real time in whole-cell extracts. We find that individual spliceosomal subcomplexes associate with pre-mRNA sequentially via an ordered pathway to yield functional spliceosomes and that association of every subcomplex is reversible. Further, early subcomplex binding events do not fully commit a pre-mRNA to splicing; rather, commitment increases as assembly proceeds. These findings have important implications for the regulation of alternative splicing. This experimental strategy should prove widely useful for mechanistic analysis of other macromolecular machines in environments approaching the complexity of living cells.
Figures
Similar articles
-
The splicing factor Prp17 interacts with the U2, U5 and U6 snRNPs and associates with the spliceosome pre- and post-catalysis.Biochem J. 2008 Dec 15;416(3):365-74. doi: 10.1042/BJ20081195. Biochem J. 2008. PMID: 18691155
-
Structure of a pre-catalytic spliceosome.Nature. 2017 Jun 29;546(7660):617-621. doi: 10.1038/nature22799. Epub 2017 May 22. Nature. 2017. PMID: 28530653 Free PMC article.
-
Prespliceosome structure provides insights into spliceosome assembly and regulation.Nature. 2018 Jul;559(7714):419-422. doi: 10.1038/s41586-018-0323-8. Epub 2018 Jul 11. Nature. 2018. PMID: 29995849 Free PMC article.
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
-
New insights into the spliceosome by single molecule fluorescence microscopy.Curr Opin Chem Biol. 2011 Dec;15(6):864-70. doi: 10.1016/j.cbpa.2011.10.010. Epub 2011 Nov 5. Curr Opin Chem Biol. 2011. PMID: 22057211 Free PMC article. Review.
Cited by
-
Second-generation covalent TMP-tag for live cell imaging.J Am Chem Soc. 2012 Aug 22;134(33):13692-9. doi: 10.1021/ja303374p. Epub 2012 Aug 9. J Am Chem Soc. 2012. PMID: 22873118 Free PMC article.
-
Pick one, but be quick: 5' splice sites and the problems of too many choices.Genes Dev. 2013 Jan 15;27(2):129-44. doi: 10.1101/gad.209759.112. Genes Dev. 2013. PMID: 23348838 Free PMC article. Review.
-
Identification of transient intermediates during spliceosome activation by single molecule fluorescence microscopy.Proc Natl Acad Sci U S A. 2022 Nov 29;119(48):e2206815119. doi: 10.1073/pnas.2206815119. Epub 2022 Nov 23. Proc Natl Acad Sci U S A. 2022. PMID: 36417433 Free PMC article.
-
Splicing Kinetics and Coordination Revealed by Direct Nascent RNA Sequencing through Nanopores.Mol Cell. 2020 Mar 5;77(5):985-998.e8. doi: 10.1016/j.molcel.2019.11.017. Epub 2019 Dec 12. Mol Cell. 2020. PMID: 31839405 Free PMC article.
-
Evaluation of fluorophores to label SNAP-tag fused proteins for multicolor single-molecule tracking microscopy in live cells.Biophys J. 2014 Aug 19;107(4):803-14. doi: 10.1016/j.bpj.2014.06.040. Biophys J. 2014. PMID: 25140415 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007596/GM/NIGMS NIH HHS/United States
- GM759628/GM/NIGMS NIH HHS/United States
- F32 GM079971/GM/NIGMS NIH HHS/United States
- R01 GM053007-15/GM/NIGMS NIH HHS/United States
- R01 GM081648-04/GM/NIGMS NIH HHS/United States
- RC1 GM091804-02/GM/NIGMS NIH HHS/United States
- R37 GM043369-21/GM/NIGMS NIH HHS/United States
- K99 GM086471-02/GM/NIGMS NIH HHS/United States
- K99/R00 GM086471/GM/NIGMS NIH HHS/United States
- GM079971/GM/NIGMS NIH HHS/United States
- R01 GM043369/GM/NIGMS NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
- F32 GM079971-03/GM/NIGMS NIH HHS/United States
- R01 GM81648/GM/NIGMS NIH HHS/United States
- R01 GM053007/GM/NIGMS NIH HHS/United States
- R37 GM043369/GM/NIGMS NIH HHS/United States
- K99 GM086471/GM/NIGMS NIH HHS/United States
- R01 GM54469/GM/NIGMS NIH HHS/United States
- T32 GM007596-30/GM/NIGMS NIH HHS/United States
- R01 GM081648/GM/NIGMS NIH HHS/United States
- RC1 GM091804/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

