Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways
- PMID: 21383145
- PMCID: PMC3064330
- DOI: 10.1073/pnas.1015312108
Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways
Abstract
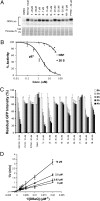
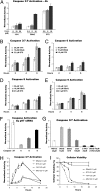
A specific small-molecule inhibitor of p97 would provide an important tool to investigate diverse functions of this essential ATPase associated with diverse cellular activities (AAA) ATPase and to evaluate its potential to be a therapeutic target in human disease. We carried out a high-throughput screen to identify inhibitors of p97 ATPase activity. Dual-reporter cell lines that simultaneously express p97-dependent and p97-independent proteasome substrates were used to stratify inhibitors that emerged from the screen. N2,N4-dibenzylquinazoline-2,4-diamine (DBeQ) was identified as a selective, potent, reversible, and ATP-competitive p97 inhibitor. DBeQ blocks multiple processes that have been shown by RNAi to depend on p97, including degradation of ubiquitin fusion degradation and endoplasmic reticulum-associated degradation pathway reporters, as well as autophagosome maturation. DBeQ also potently inhibits cancer cell growth and is more rapid than a proteasome inhibitor at mobilizing the executioner caspases-3 and -7. Our results provide a rationale for targeting p97 in cancer therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Development of p97 AAA ATPase inhibitors.Autophagy. 2011 Sep;7(9):1091-2. doi: 10.4161/auto.7.9.16489. Epub 2011 Sep 1. Autophagy. 2011. PMID: 21606684 Free PMC article.
-
Structure-activity relationship study reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase.ChemMedChem. 2013 Feb;8(2):297-312. doi: 10.1002/cmdc.201200520. Epub 2013 Jan 11. ChemMedChem. 2013. PMID: 23316025 Free PMC article.
-
Novel p97/VCP inhibitor induces endoplasmic reticulum stress and apoptosis in both bortezomib-sensitive and -resistant multiple myeloma cells.Cancer Sci. 2019 Oct;110(10):3275-3287. doi: 10.1111/cas.14154. Epub 2019 Aug 14. Cancer Sci. 2019. PMID: 31368616 Free PMC article.
-
Structure and Function of the AAA+ ATPase p97, a Key Player in Protein Homeostasis.Subcell Biochem. 2019;93:221-272. doi: 10.1007/978-3-030-28151-9_7. Subcell Biochem. 2019. PMID: 31939153 Review.
-
The AAA+ ATPase p97, a cellular multitool.Biochem J. 2017 Aug 17;474(17):2953-2976. doi: 10.1042/BCJ20160783. Biochem J. 2017. PMID: 28819009 Free PMC article. Review.
Cited by
-
An Integrative Synthetic Biology Approach to Interrogating Cellular Ubiquitin and Ufm Signaling.Int J Mol Sci. 2020 Jun 14;21(12):4231. doi: 10.3390/ijms21124231. Int J Mol Sci. 2020. PMID: 32545848 Free PMC article.
-
mTOR dysfunction contributes to vacuolar pathology and weakness in valosin-containing protein associated inclusion body myopathy.Hum Mol Genet. 2013 Mar 15;22(6):1167-79. doi: 10.1093/hmg/dds524. Epub 2012 Dec 18. Hum Mol Genet. 2013. PMID: 23250913 Free PMC article.
-
White Spot Syndrome Virus Benefits from Endosomal Trafficking, Substantially Facilitated by a Valosin-Containing Protein, To Escape Autophagic Elimination and Propagate in the Crustacean Cherax quadricarinatus.J Virol. 2020 Nov 23;94(24):e01570-20. doi: 10.1128/JVI.01570-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967962 Free PMC article.
-
The coordinated action of VCP/p97 and GCN2 regulates cancer cell metabolism and proteostasis during nutrient limitation.Oncogene. 2019 Apr;38(17):3216-3231. doi: 10.1038/s41388-018-0651-z. Epub 2019 Jan 9. Oncogene. 2019. PMID: 30626938 Free PMC article.
-
Host AAA+ ATPase TER94 Plays Critical Roles in Building the Baculovirus Viral Replication Factory and Virion Morphogenesis.J Virol. 2020 Feb 28;94(6):e01674-19. doi: 10.1128/JVI.01674-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896597 Free PMC article.
References
-
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. - PubMed
-
- Müller JM, Deinhardt K, Rosewell I, Warren G, Shima DT. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem Biophys Res Commun. 2007;354:459–465. - PubMed
-
- Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J. The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem. 1999;380:1049–1062. - PubMed
-
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

