The structural basis for MCM2-7 helicase activation by GINS and Cdc45
- PMID: 21378962
- PMCID: PMC4184033
- DOI: 10.1038/nsmb.2004
The structural basis for MCM2-7 helicase activation by GINS and Cdc45
Abstract
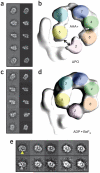
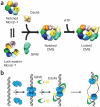
Two central steps for initiating eukaryotic DNA replication involve loading of the Mcm2-7 helicase onto double-stranded DNA and its activation by GINS-Cdc45. To better understand these events, we determined the structures of Mcm2-7 and the CMG complex by using single-particle electron microscopy. Mcm2-7 adopts two conformations--a lock-washer-shaped spiral state and a planar, gapped-ring form--in which Mcm2 and Mcm5 flank a breach in the helicase perimeter. GINS and Cdc45 bridge this gap, forming a topologically closed assembly with a large interior channel; nucleotide binding further seals off the discontinuity between Mcm2 and Mcm5, partitioning the channel into two smaller pores. Together, our data help explain how GINS and Cdc45 activate Mcm2-7, indicate that Mcm2-7 loading may be assisted by a natural predisposition of the hexamer to form open rings, and suggest a mechanism by which the CMG complex assists DNA strand separation.
Figures




Similar articles
-
DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome.Elife. 2014 Aug 12;3:e03273. doi: 10.7554/eLife.03273. Elife. 2014. PMID: 25117490 Free PMC article.
-
Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement.Proc Natl Acad Sci U S A. 2015 Jan 20;112(3):E249-58. doi: 10.1073/pnas.1422003112. Epub 2015 Jan 5. Proc Natl Acad Sci U S A. 2015. PMID: 25561522 Free PMC article.
-
DDK regulates replication initiation by controlling the multiplicity of Cdc45-GINS binding to Mcm2-7.Elife. 2021 Feb 22;10:e65471. doi: 10.7554/eLife.65471. Elife. 2021. PMID: 33616038 Free PMC article.
-
The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review.
-
The eukaryotic Mcm2-7 replicative helicase.Subcell Biochem. 2012;62:113-34. doi: 10.1007/978-94-007-4572-8_7. Subcell Biochem. 2012. PMID: 22918583 Review.
Cited by
-
Structure and evolutionary origins of the CMG complex.Chromosoma. 2013 Mar;122(1-2):47-53. doi: 10.1007/s00412-013-0397-x. Epub 2013 Feb 15. Chromosoma. 2013. PMID: 23412083 Review.
-
Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components.EMBO J. 2012 May 2;31(9):2182-94. doi: 10.1038/emboj.2012.68. Epub 2012 Mar 20. EMBO J. 2012. PMID: 22433840 Free PMC article.
-
In Vitro Reconstitution Defines the Minimal Requirements for Cdc48-Dependent Disassembly of the CMG Helicase in Budding Yeast.Cell Rep. 2019 Sep 10;28(11):2777-2783.e4. doi: 10.1016/j.celrep.2019.08.026. Cell Rep. 2019. PMID: 31509741 Free PMC article.
-
Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.Mol Cell. 2019 Apr 4;74(1):173-184.e4. doi: 10.1016/j.molcel.2019.01.023. Epub 2019 Feb 20. Mol Cell. 2019. PMID: 30797687 Free PMC article.
-
CMG helicase can use ATPγS to unwind DNA: Implications for the rate-limiting step in the reaction mechanism.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2119580119. doi: 10.1073/pnas.2119580119. Proc Natl Acad Sci U S A. 2022. PMID: 35042821 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

