G4 motifs correlate with promoter-proximal transcriptional pausing in human genes
- PMID: 21371997
- PMCID: PMC3130262
- DOI: 10.1093/nar/gkr079
G4 motifs correlate with promoter-proximal transcriptional pausing in human genes
Abstract
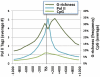
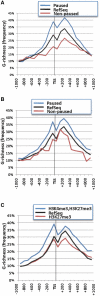
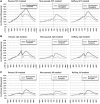
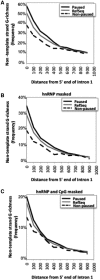
The RNA Pol II transcription complex pauses just downstream of the promoter in a significant fraction of human genes. The local features of genomic structure that contribute to pausing have not been defined. Here, we show that genes that pause are more G-rich within the region flanking the transcription start site (TSS) than RefSeq genes or non-paused genes. We show that enrichment of binding motifs for common transcription factors, such as SP1, may account for G-richness upstream but not downstream of the TSS. We further show that pausing correlates with the presence of a GrIn1 element, an element bearing one or more G4 motifs at the 5'-end of the first intron, on the non-template DNA strand. These results suggest potential roles for dynamic G4 DNA and G4 RNA structures in cis-regulation of pausing, and thus genome-wide regulation of gene expression, in human cells.
Figures

Similar articles
-
Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes.Nucleic Acids Res. 2008 Mar;36(4):1321-33. doi: 10.1093/nar/gkm1138. Epub 2008 Jan 10. Nucleic Acids Res. 2008. PMID: 18187510 Free PMC article.
-
Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters.Genomics. 2004 May;83(5):873-82. doi: 10.1016/j.ygeno.2003.11.001. Genomics. 2004. PMID: 15081116
-
Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation.Genes Dev. 1995 Mar 1;9(5):559-72. doi: 10.1101/gad.9.5.559. Genes Dev. 1995. PMID: 7698646
-
Regulation of c-myc and immunoglobulin kappa gene transcription by promoter-proximal pausing of RNA polymerase II.Curr Top Microbiol Immunol. 1999;246:225-31. doi: 10.1007/978-3-642-60162-0_28. Curr Top Microbiol Immunol. 1999. PMID: 10396060 Review.
-
Pause & go: from the discovery of RNA polymerase pausing to its functional implications.Curr Opin Cell Biol. 2017 Jun;46:72-80. doi: 10.1016/j.ceb.2017.03.002. Epub 2017 Mar 28. Curr Opin Cell Biol. 2017. PMID: 28363125 Free PMC article. Review.
Cited by
-
Exploring possible DNA structures in real-time polymerase kinetics using Pacific Biosciences sequencer data.BMC Bioinformatics. 2015 Jan 28;16:21. doi: 10.1186/s12859-014-0449-0. BMC Bioinformatics. 2015. PMID: 25626999 Free PMC article.
-
Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop 'G4 Kissing' interaction.Nucleic Acids Res. 2020 Jun 19;48(11):5907-5925. doi: 10.1093/nar/gkaa357. Nucleic Acids Res. 2020. PMID: 32383760 Free PMC article.
-
Double-stranded flanking ends affect the folding kinetics and conformational equilibrium of G-quadruplexes forming sequences within the promoter of KIT oncogene.Nucleic Acids Res. 2021 Sep 27;49(17):9724-9737. doi: 10.1093/nar/gkab674. Nucleic Acids Res. 2021. PMID: 34478543 Free PMC article.
-
The Human Integrator Complex Facilitates Transcriptional Elongation by Endonucleolytic Cleavage of Nascent Transcripts.Cell Rep. 2020 Jul 21;32(3):107917. doi: 10.1016/j.celrep.2020.107917. Cell Rep. 2020. PMID: 32697989 Free PMC article.
-
Strand invasion of DNA quadruplexes by PNA: comparison of homologous and complementary hybridization.Chembiochem. 2013 Aug 19;14(12):1476-84. doi: 10.1002/cbic.201300263. Epub 2013 Jul 19. Chembiochem. 2013. PMID: 23868291 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. - PubMed
-
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

