Protein folding at the exit tunnel
- PMID: 21370971
- PMCID: PMC5807062
- DOI: 10.1146/annurev-biophys-042910-155338
Protein folding at the exit tunnel
Abstract
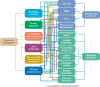
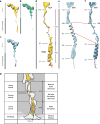
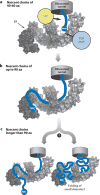
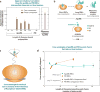
Over five decades of research have yielded a large body of information on how purified proteins attain their native state when refolded in the test tube, starting from a chemically or thermally denatured state. Nevertheless, we still know little about how proteins fold and unfold in their natural biological habitat: the living cell. Indeed, a variety of cellular components, including molecular chaperones, the ribosome, and crowding of the intracellular medium, modulate folding mechanisms in physiologically relevant environments. This review focuses on the current state of knowledge in protein folding in the cell with emphasis on the early stage of a protein's life, as the nascent polypeptide traverses and emerges from the ribosomal tunnel. Given the vectorial nature of ribosome-assisted translation, the transient degree of chain elongation becomes a relevant variable expected to affect nascent protein foldability, aggregation propensity and extent of interaction with chaperones and the ribosome.
Figures


Similar articles
-
Role of the ribosome in protein folding.Biotechnol J. 2008 Aug;3(8):999-1009. doi: 10.1002/biot.200800098. Biotechnol J. 2008. PMID: 18702035
-
Residue-specific contact order and contact breadth in single-domain proteins: implications for folding as a function of chain elongation.Biotechnol Prog. 2008 May-Jun;24(3):570-5. doi: 10.1021/bp070475v. Epub 2008 May 10. Biotechnol Prog. 2008. PMID: 18471028
-
alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel.Nat Struct Mol Biol. 2010 Mar;17(3):313-7. doi: 10.1038/nsmb.1756. Epub 2010 Feb 7. Nat Struct Mol Biol. 2010. PMID: 20139981
-
Cotranslational Folding of Proteins on the Ribosome.Biomolecules. 2020 Jan 7;10(1):97. doi: 10.3390/biom10010097. Biomolecules. 2020. PMID: 31936054 Free PMC article. Review.
-
Mechanisms of protein folding.Eur Biophys J. 2008 Jul;37(6):721-8. doi: 10.1007/s00249-007-0256-x. Epub 2008 Jan 9. Eur Biophys J. 2008. PMID: 18185928 Review.
Cited by
-
Increased ribosome density associated to positively charged residues is evident in ribosome profiling experiments performed in the absence of translation inhibitors.RNA Biol. 2016 Jun 2;13(6):561-8. doi: 10.1080/15476286.2016.1172755. Epub 2016 Apr 11. RNA Biol. 2016. PMID: 27064519 Free PMC article.
-
Solid-state NMR enhanced by dynamic nuclear polarization as a novel tool for ribosome structural biology.J Biomol NMR. 2013 Jun;56(2):85-93. doi: 10.1007/s10858-013-9721-2. Epub 2013 May 21. J Biomol NMR. 2013. PMID: 23689811
-
Rqc1 and other yeast proteins containing highly positively charged sequences are not targets of the RQC complex.J Biol Chem. 2021 Jan-Jun;296:100586. doi: 10.1016/j.jbc.2021.100586. Epub 2021 Mar 24. J Biol Chem. 2021. PMID: 33774050 Free PMC article.
-
Site-specific fluorescent labeling of nascent proteins on the translating ribosome.J Am Chem Soc. 2011 Sep 28;133(38):14936-9. doi: 10.1021/ja206626g. Epub 2011 Sep 2. J Am Chem Soc. 2011. PMID: 21870811 Free PMC article.
-
Does mRNA structure contain genetic information for regulating co-translational protein folding?Zool Res. 2017 Jan 18;38(1):36-43. doi: 10.13918/j.issn.2095-8137.2017.004. Zool Res. 2017. PMID: 28271668 Free PMC article. Review.
References
-
- Anfinsen CB, Redfield RR, Choate WI, Page J, Carroll WR. Studies on the gross structure, cross-linkages, and terminal sequences in ribonuclease. J Biol Chem. 1954;207:201–10. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–20. - PubMed
-
- Bhushan S, Gartmann M, Halic M, Armache J-P, Jarasch A, et al. Alpha-helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17:313–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

