Mass spectrometry-based immuno-precipitation proteomics - the user's guide
- PMID: 21365760
- PMCID: PMC3708439
- DOI: 10.1002/pmic.201000548
Mass spectrometry-based immuno-precipitation proteomics - the user's guide
Abstract
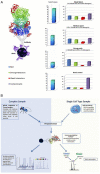
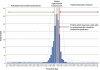
Immuno-precipitation (IP) experiments using MS provide a sensitive and accurate way of characterising protein complexes and their response to regulatory mechanisms. Differences in stoichiometry can be determined as well as the reliable identification of specific binding partners. The quality control of IP and protein interaction studies has its basis in the biology that is being observed. Is that unusual protein identification a genuine novelty, or an experimental irregularity? Antibodies and the solid matrices used in these techniques isolate not only the target protein and its specific interaction partners but also many non-specific 'contaminants' requiring a structured analysis strategy. These methodological developments and the speed and accuracy of MS machines, which has been increasing consistently in the last 5 years, have expanded the number of proteins identified and complexity of analysis. The European Science Foundation's Frontiers in Functional Genomics programme 'Quality Control in Proteomics' Workshop provided a forum for disseminating knowledge and experience on this subject. Our aim in this technical brief is to outline clearly, for the scientists wanting to carry out this kind of experiment, and recommend what, in our experience, are the best potential ways to design an IP experiment, to help identify possible pitfalls, discuss important controls and outline how to manage and analyse the large amount of data generated. Detailed experimental methodologies have been referenced but not described in the form of protocols.
Copyright © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

Similar articles
-
ROCS: a reproducibility index and confidence score for interaction proteomics studies.BMC Bioinformatics. 2012 Jun 8;13:128. doi: 10.1186/1471-2105-13-128. BMC Bioinformatics. 2012. PMID: 22682516 Free PMC article.
-
Quality control in mass spectrometry-based proteomics.Mass Spectrom Rev. 2018 Sep;37(5):697-711. doi: 10.1002/mas.21544. Epub 2017 Sep 7. Mass Spectrom Rev. 2018. PMID: 28802010 Review.
-
Identification of Plant Chromatin Interaction Networks Using IP-MS and co-IP.Methods Mol Biol. 2025;2873:129-143. doi: 10.1007/978-1-0716-4228-3_8. Methods Mol Biol. 2025. PMID: 39576600
-
Recovering protein-protein and domain-domain interactions from aggregation of IP-MS proteomics of coregulator complexes.PLoS Comput Biol. 2011 Dec;7(12):e1002319. doi: 10.1371/journal.pcbi.1002319. Epub 2011 Dec 29. PLoS Comput Biol. 2011. PMID: 22219718 Free PMC article.
-
Identifying components of protein complexes in C. elegans using co-immunoprecipitation and mass spectrometry.J Proteomics. 2010 Oct 10;73(11):2198-204. doi: 10.1016/j.jprot.2010.05.008. Epub 2010 May 28. J Proteomics. 2010. PMID: 20546956 Free PMC article. Review.
Cited by
-
Studying Translesion DNA Synthesis Using Xenopus In Vitro Systems.Methods Mol Biol. 2024;2740:21-36. doi: 10.1007/978-1-0716-3557-5_2. Methods Mol Biol. 2024. PMID: 38393467
-
Identification of cellular proteins interacting with PEDV M protein through APEX2 labeling.J Proteomics. 2021 May 30;240:104191. doi: 10.1016/j.jprot.2021.104191. Epub 2021 Mar 20. J Proteomics. 2021. PMID: 33757879 Free PMC article.
-
Fluid Candidate Biomarkers for Alzheimer's Disease: A Precision Medicine Approach.J Pers Med. 2020 Nov 11;10(4):221. doi: 10.3390/jpm10040221. J Pers Med. 2020. PMID: 33187336 Free PMC article. Review.
-
Cell Cycle-Specific Protein Phosphatase 1 (PP1) Substrates Identification Using Genetically Modified Cell Lines.Methods Mol Biol. 2024;2740:37-61. doi: 10.1007/978-1-0716-3557-5_3. Methods Mol Biol. 2024. PMID: 38393468
-
Identification of new binding proteins of focal adhesion kinase using immunoprecipitation and mass spectrometry.Sci Rep. 2019 Sep 9;9(1):12908. doi: 10.1038/s41598-019-49145-6. Sci Rep. 2019. PMID: 31501460 Free PMC article.
References
-
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. - PubMed
-
- Burnette W. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981;112:195–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 097945/WT_/Wellcome Trust/United Kingdom
- G0301131/MRC_/Medical Research Council/United Kingdom
- 073980/WT_/Wellcome Trust/United Kingdom
- 083524/WT_/Wellcome Trust/United Kingdom
- 081361/WT_/Wellcome Trust/United Kingdom
- G0801738/MRC_/Medical Research Council/United Kingdom
- 037538/WT_/Wellcome Trust/United Kingdom
- 073980/Z/03/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- C12944/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- C08577/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

