Prediction of Associations between microRNAs and Gene Expression in Glioma Biology
- PMID: 21358821
- PMCID: PMC3040173
- DOI: 10.1371/journal.pone.0014681
Prediction of Associations between microRNAs and Gene Expression in Glioma Biology
Abstract
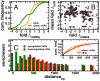
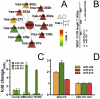
Despite progress in the determination of miR interactions, their regulatory role in cancer is only beginning to be unraveled. Utilizing gene expression data from 27 glioblastoma samples we found that the mere knowledge of physical interactions between specific mRNAs and miRs can be used to determine associated regulatory interactions, allowing us to identify 626 associated interactions, involving 128 miRs that putatively modulate the expression of 246 mRNAs. Experimentally determining the expression of miRs, we found an over-representation of over(under)-expressed miRs with various predicted mRNA target sequences. Such significantly associated miRs that putatively bind over-expressed genes strongly tend to have binding sites nearby the 3'UTR of the corresponding mRNAs, suggesting that the presence of the miRs near the translation stop site may be a factor in their regulatory ability. Our analysis predicted a significant association between miR-128 and the protein kinase WEE1, which we subsequently validated experimentally by showing that the over-expression of the naturally under-expressed miR-128 in glioma cells resulted in the inhibition of WEE1 in glioblastoma cells.
Conflict of interest statement
Figures



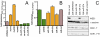
Similar articles
-
Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas.J Clin Endocrinol Metab. 2010 Oct;95(10):E181-91. doi: 10.1210/jc.2010-0581. Epub 2010 Jul 28. J Clin Endocrinol Metab. 2010. PMID: 20668041
-
Regulation of IL-8 gene expression in gliomas by microRNA miR-93.BMC Cancer. 2015 Oct 8;15:661. doi: 10.1186/s12885-015-1659-1. BMC Cancer. 2015. PMID: 26449498 Free PMC article.
-
Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas.Oncotarget. 2017 Feb 7;8(6):10287-10297. doi: 10.18632/oncotarget.14396. Oncotarget. 2017. PMID: 28055966 Free PMC article.
-
Whole-Genome Expression Microarray Combined with Machine Learning to Identify Prognostic Biomarkers for High-Grade Glioma.J Mol Neurosci. 2018 Apr;64(4):491-500. doi: 10.1007/s12031-018-1049-7. Epub 2018 Mar 3. J Mol Neurosci. 2018. PMID: 29502292
-
Long non-coding RNAs (lncRNAs) and microRNAs regulatory pathways in the tumorigenesis and pathogenesis of glioma.Discov Med. 2019 Sep;28(153):129-138. Discov Med. 2019. PMID: 31926584 Review.
Cited by
-
Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma.Neuro Oncol. 2012 Sep;14(9):1153-62. doi: 10.1093/neuonc/nos145. Epub 2012 Jul 27. Neuro Oncol. 2012. PMID: 22844109 Free PMC article.
-
Synthetic and Medicinal Chemistry Approaches Toward WEE1 Kinase Inhibitors and Its Degraders.ACS Omega. 2023 Jun 2;8(23):20196-20233. doi: 10.1021/acsomega.3c01558. eCollection 2023 Jun 13. ACS Omega. 2023. PMID: 37323408 Free PMC article. Review.
-
Intervention in prediction measure: a new approach to assessing variable importance for random forests.BMC Bioinformatics. 2017 May 2;18(1):230. doi: 10.1186/s12859-017-1650-8. BMC Bioinformatics. 2017. PMID: 28464827 Free PMC article.
-
Uncovering MicroRNA and Transcription Factor Mediated Regulatory Networks in Glioblastoma.PLoS Comput Biol. 2012;8(7):e1002488. doi: 10.1371/journal.pcbi.1002488. Epub 2012 Jul 19. PLoS Comput Biol. 2012. PMID: 22829753 Free PMC article.
-
The Emerging Field of Noncoding RNAs and Their Importance in Pediatric Diseases.J Pediatr. 2020 Jun;221S:S11-S19. doi: 10.1016/j.jpeds.2020.02.078. J Pediatr. 2020. PMID: 32482229 Free PMC article. No abstract available.
References
-
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. - PubMed
-
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. - PubMed
-
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. - PubMed
-
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

