NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly
- PMID: 21347350
- PMCID: PMC3037360
- DOI: 10.1371/journal.ppat.1001278
NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly
Abstract
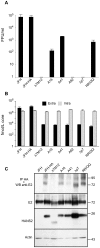
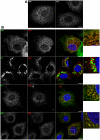
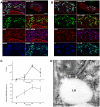
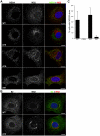
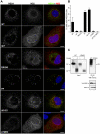
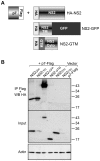
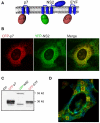
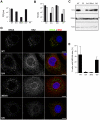
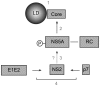
Growing experimental evidence indicates that, in addition to the physical virion components, the non-structural proteins of hepatitis C virus (HCV) are intimately involved in orchestrating morphogenesis. Since it is dispensable for HCV RNA replication, the non-structural viral protein NS2 is suggested to play a central role in HCV particle assembly. However, despite genetic evidences, we have almost no understanding about NS2 protein-protein interactions and their role in the production of infectious particles. Here, we used co-immunoprecipitation and/or fluorescence resonance energy transfer with fluorescence lifetime imaging microscopy analyses to study the interactions between NS2 and the viroporin p7 and the HCV glycoprotein E2. In addition, we used alanine scanning insertion mutagenesis as well as other mutations in the context of an infectious virus to investigate the functional role of NS2 in HCV assembly. Finally, the subcellular localization of NS2 and several mutants was analyzed by confocal microscopy. Our data demonstrate molecular interactions between NS2 and p7 and E2. Furthermore, we show that, in the context of an infectious virus, NS2 accumulates over time in endoplasmic reticulum-derived dotted structures and colocalizes with both the envelope glycoproteins and components of the replication complex in close proximity to the HCV core protein and lipid droplets, a location that has been shown to be essential for virus assembly. We show that NS2 transmembrane region is crucial for both E2 interaction and subcellular localization. Moreover, specific mutations in core, envelope proteins, p7 and NS5A reported to abolish viral assembly changed the subcellular localization of NS2 protein. Together, these observations indicate that NS2 protein attracts the envelope proteins at the assembly site and it crosstalks with non-structural proteins for virus assembly.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Detergent-resistant membrane association of NS2 and E2 during hepatitis C virus replication.J Virol. 2015 Apr;89(8):4562-74. doi: 10.1128/JVI.00123-15. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673706 Free PMC article.
-
Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins.J Virol. 2011 Jan;85(1):86-97. doi: 10.1128/JVI.01070-10. Epub 2010 Oct 20. J Virol. 2011. PMID: 20962101 Free PMC article.
-
The amino-terminus of the hepatitis C virus (HCV) p7 viroporin and its cleavage from glycoprotein E2-p7 precursor determine specific infectivity and secretion levels of HCV particle types.PLoS Pathog. 2017 Dec 18;13(12):e1006774. doi: 10.1371/journal.ppat.1006774. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29253880 Free PMC article.
-
Hepatitis C virus proteins: from structure to function.Curr Top Microbiol Immunol. 2013;369:113-42. doi: 10.1007/978-3-642-27340-7_5. Curr Top Microbiol Immunol. 2013. PMID: 23463199 Review.
-
Hepatitis C virus: viral proteins on the move.Biochem Soc Trans. 2009 Oct;37(Pt 5):986-90. doi: 10.1042/BST0370986. Biochem Soc Trans. 2009. PMID: 19754437 Review.
Cited by
-
Cell-death-inducing DFFA-like Effector B Contributes to the Assembly of Hepatitis C Virus (HCV) Particles and Interacts with HCV NS5A.Sci Rep. 2016 Jun 10;6:27778. doi: 10.1038/srep27778. Sci Rep. 2016. PMID: 27282740 Free PMC article.
-
The acidic domain of the hepatitis C virus NS4A protein is required for viral assembly and envelopment through interactions with the viral E1 glycoprotein.PLoS Pathog. 2019 Feb 7;15(2):e1007163. doi: 10.1371/journal.ppat.1007163. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30730994 Free PMC article.
-
NS2 proteins of GB virus B and hepatitis C virus share common protease activities and membrane topologies.J Virol. 2014 Jul;88(13):7426-44. doi: 10.1128/JVI.00656-14. Epub 2014 Apr 16. J Virol. 2014. PMID: 24741107 Free PMC article.
-
Genetic and functional characterization of the N-terminal region of the hepatitis C virus NS2 protein.J Virol. 2013 Apr;87(8):4130-45. doi: 10.1128/JVI.03174-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408609 Free PMC article.
-
Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion.J Virol. 2016 Jul 27;90(16):7159-70. doi: 10.1128/JVI.00826-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27226379 Free PMC article.
References
-
- Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. - PubMed
-
- Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. - PubMed
-
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia, Pa: Lippincott Williams & Wilkins; 2007. pp. 1101–1152.
-
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

