Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens
- PMID: 21345963
- PMCID: PMC3126233
- DOI: 10.1128/JVI.02077-10
Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens
Abstract
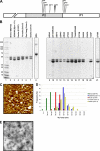
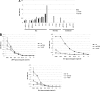
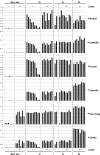
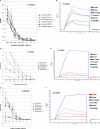
Noroviruses (NoVs) are one of the leading causes of gastroenteritis in children and adults. For the last 2 decades, genogroup II genotype 4 (GII.4) NoVs have been circulating worldwide. GII.4 NoVs can be divided into variants, and since 2002 they have circulated in the population before being replaced every 2 or 3 years, which raises questions about the role of their histo-blood group antigen (HBGA) ligands in their evolution. To shed light on these questions, we performed an analysis of the interaction between representative GII.4 variants and HBGAs, and we determined the role of selected amino acids in the binding profiles. By mutagenesis, we showed that there was a strict structural requirement for the amino acids, directly implicated in interactions with HBGAs. However, the ablation of the threonine residue at position 395 (ΔT395), an epidemiological feature of the post-2002 variants, was not deleterious to the binding of the virus-like particle (VLP) to the H antigen, while binding to A and B antigens was severely hampered. Nevertheless, the ΔT395 VLPs gained the capacity to bind to the Lewis x and sialyl-Lewis x antigens in comparison with the wild-type VLP, demonstrating that amino acid residues outside the HBGA binding site can modify the binding properties of NoVs. We also analyzed the attachment of baculovirus-expressed VLPs from six variants (Bristol, US95/96, Hunter, Yerseke, Den Haag, and Osaka) that were isolated from 1987 to 2007 to phenotyped saliva samples and synthetic HBGAs. We showed that the six variants could all attach to saliva of secretors irrespective of the ABO phenotype and to oligosaccharides characteristic of the secretor phenotype. Interestingly, Den Haag and Osaka variants additionally bound to carbohydrates present in the saliva of Lewis-positive nonsecretors. The carbohydrate binding profile and the genetic and mutagenesis analysis suggested that GII.4 binding to Lewis x and sialyl-Lewis x antigens might be a by-product of the genetic variation of the amino acids located in the vicinity of the binding site. Analysis of the binding properties for the six variants by surface plasmon resonance showed that only post-2002 variants (i.e., Hunter, Yerseke, Den Haag, and Osaka) presented strong binding to A and B antigens, suggesting that the GII.4 evolution could be related to an increased affinity for HBGAs for the post-2002 variants. The combination of increased affinity for ABH antigens and of a newly acquired ability to recognize glycans from Lewis-positive nonsecretors could have contributed to the epidemiological importance of strains such as the Den Haag GII.4 subtype.
Figures

Similar articles
-
Noroviruses distinguish between type 1 and type 2 histo-blood group antigens for binding.J Virol. 2008 Nov;82(21):10756-67. doi: 10.1128/JVI.00802-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701592 Free PMC article.
-
Antibodies against Lewis antigens inhibit the binding of human norovirus GII.4 virus-like particles to saliva but not to intestinal Caco-2 cells.Virol J. 2016 May 21;13:82. doi: 10.1186/s12985-016-0538-y. Virol J. 2016. PMID: 27206610 Free PMC article.
-
Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.J Virol. 2014 Jun;88(11):6168-80. doi: 10.1128/JVI.00201-14. Epub 2014 Mar 19. J Virol. 2014. PMID: 24648450 Free PMC article.
-
Norovirus and histo-blood group antigens.Jpn J Infect Dis. 2011;64(2):95-103. Jpn J Infect Dis. 2011. PMID: 21519121 Review.
-
Histo-blood group antigens: a common niche for norovirus and rotavirus.Expert Rev Mol Med. 2014 Mar 10;16:e5. doi: 10.1017/erm.2014.2. Expert Rev Mol Med. 2014. PMID: 24606759 Review.
Cited by
-
Functional and structural characterization of Norovirus GII.6 in recognizing histo-blood group antigens.Virol Sin. 2023 Feb;38(1):56-65. doi: 10.1016/j.virs.2022.09.010. Epub 2022 Oct 7. Virol Sin. 2023. PMID: 36216242 Free PMC article.
-
Glycan Recognition in Human Norovirus Infections.Viruses. 2021 Oct 14;13(10):2066. doi: 10.3390/v13102066. Viruses. 2021. PMID: 34696500 Free PMC article. Review.
-
Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens.PLoS Pathog. 2011 Jul;7(7):e1002152. doi: 10.1371/journal.ppat.1002152. Epub 2011 Jul 21. PLoS Pathog. 2011. PMID: 21811409 Free PMC article.
-
Monoclonal Antibodies Generated against Glycoconjugates Recognize Chemical Linkers.Antibodies (Basel). 2020 Sep 15;9(3):48. doi: 10.3390/antib9030048. Antibodies (Basel). 2020. PMID: 32942538 Free PMC article.
-
Intrahost Norovirus Evolution in Chronic Infection Over 5 Years of Shedding in a Kidney Transplant Recipient.Front Microbiol. 2018 Mar 2;9:371. doi: 10.3389/fmicb.2018.00371. eCollection 2018. Front Microbiol. 2018. PMID: 29552005 Free PMC article.
References
-
- Boireau W., Rouleau A., Lucchi G., Ducoroy P. 2009. Revisited BIA-MS combination: entire “on-a-chip” processing leading to the proteins identification at low femtomole to sub-femtomole levels. Biosens. Bioelectron. 24:1121–1127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

