Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus
- PMID: 21336280
- PMCID: PMC3103780
- DOI: 10.1038/ng.766
Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus
Abstract
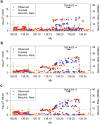
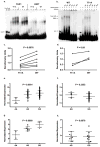
Systemic lupus erythematosus (SLE, MIM152700) is an autoimmune disease characterized by self-reactive antibodies resulting in systemic inflammation and organ failure. TNFAIP3, encoding the ubiquitin-modifying enzyme A20, is an established susceptibility locus for SLE. By fine mapping and genomic re-sequencing in ethnically diverse populations, we fully characterized the TNFAIP3 risk haplotype and identified a TT>A polymorphic dinucleotide (deletion T followed by a T to A transversion) associated with SLE in subjects of European (P = 1.58 × 10(-8), odds ratio = 1.70) and Korean (P = 8.33 × 10(-10), odds ratio = 2.54) ancestry. This variant, located in a region of high conservation and regulatory potential, bound a nuclear protein complex composed of NF-κB subunits with reduced avidity. Further, compared with the non-risk haplotype, the haplotype carrying this variant resulted in reduced TNFAIP3 mRNA and A20 protein expression. These results establish this TT>A variant as the most likely functional polymorphism responsible for the association between TNFAIP3 and SLE.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression.PLoS Genet. 2013;9(9):e1003750. doi: 10.1371/journal.pgen.1003750. Epub 2013 Sep 5. PLoS Genet. 2013. PMID: 24039598 Free PMC article.
-
Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus.Nat Genet. 2008 Sep;40(9):1062-4. doi: 10.1038/ng.202. Nat Genet. 2008. PMID: 19165919 Free PMC article.
-
The association between BANK1 and TNFAIP3 gene polymorphisms and systemic lupus erythematosus: a meta-analysis.Int J Immunogenet. 2011 Apr;38(2):151-9. doi: 10.1111/j.1744-313X.2010.00990.x. Epub 2011 Jan 5. Int J Immunogenet. 2011. PMID: 21208380
-
Single nucleotide polymorphisms at the TNFAIP3/A20 locus and susceptibility/resistance to inflammatory and autoimmune diseases.Adv Exp Med Biol. 2014;809:163-83. doi: 10.1007/978-1-4939-0398-6_10. Adv Exp Med Biol. 2014. PMID: 25302371 Review.
-
Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease.Biochem Soc Trans. 2011 Aug;39(4):1086-91. doi: 10.1042/BST0391086. Biochem Soc Trans. 2011. PMID: 21787353 Review.
Cited by
-
Immune-regulatory mechanisms in systemic autoimmune and rheumatic diseases.Clin Dev Immunol. 2012;2012:941346. doi: 10.1155/2012/941346. Epub 2011 Oct 27. Clin Dev Immunol. 2012. PMID: 22110541 Free PMC article. Review.
-
The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis.Nat Immunol. 2015 Jun;16(6):618-27. doi: 10.1038/ni.3172. Epub 2015 May 4. Nat Immunol. 2015. PMID: 25939025 Free PMC article.
-
Emerging therapies for systemic lupus erythematosus--focus on targeting interferon-alpha.Clin Immunol. 2012 Jun;143(3):210-21. doi: 10.1016/j.clim.2012.03.005. Epub 2012 Apr 6. Clin Immunol. 2012. PMID: 22525889 Free PMC article. Review.
-
Recent insights into the genetic basis of systemic lupus erythematosus.Ann Rheum Dis. 2013 Apr;72 Suppl 2(0 2):ii56-61. doi: 10.1136/annrheumdis-2012-202351. Epub 2012 Dec 19. Ann Rheum Dis. 2013. PMID: 23253915 Free PMC article. Review.
-
Integrative omics for health and disease.Nat Rev Genet. 2018 May;19(5):299-310. doi: 10.1038/nrg.2018.4. Epub 2018 Feb 26. Nat Rev Genet. 2018. PMID: 29479082 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 AR055385/AR/NIAMS NIH HHS/United States
- RC1 AR058621-01/AR/NIAMS NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- UL1 RR025741/RR/NCRR NIH HHS/United States
- R01 AR056360-01A1/AR/NIAMS NIH HHS/United States
- R01AR051545-01A2/AR/NIAMS NIH HHS/United States
- P30 AR053483-01A1/AR/NIAMS NIH HHS/United States
- K24 AI078004/AI/NIAID NIH HHS/United States
- P20 RR020143-01/RR/NCRR NIH HHS/United States
- P60 AR053308/AR/NIAMS NIH HHS/United States
- N01AR62277/AR/NIAMS NIH HHS/United States
- R01 AI063274/AI/NIAID NIH HHS/United States
- M01 RR000079/RR/NCRR NIH HHS/United States
- R01 AR043274-11/AR/NIAMS NIH HHS/United States
- R01 DE018209/DE/NIDCR NIH HHS/United States
- P01 AR49084/AR/NIAMS NIH HHS/United States
- P20 RR020143/RR/NCRR NIH HHS/United States
- P30 GM103510/GM/NIGMS NIH HHS/United States
- P60 AR053308-01/AR/NIAMS NIH HHS/United States
- R01 AR043727-10A1/AR/NIAMS NIH HHS/United States
- UL1 RR024999/RR/NCRR NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- R01 AR043814-13/AR/NIAMS NIH HHS/United States
- M01 RR-00079/RR/NCRR NIH HHS/United States
- K08 AI083790/AI/NIAID NIH HHS/United States
- UL1 TR000154/TR/NCATS NIH HHS/United States
- R01 AR033062-17/AR/NIAMS NIH HHS/United States
- P20 RR015577/RR/NCRR NIH HHS/United States
- P30 RR031152/RR/NCRR NIH HHS/United States
- P20 RR015577-08/RR/NCRR NIH HHS/United States
- R01 DE018209-01A1/DE/NIDCR NIH HHS/United States
- R01 AR042460-06A1/AR/NIAMS NIH HHS/United States
- N01 AR062277/AR/NIAMS NIH HHS/United States
- R01 AR056360/AR/NIAMS NIH HHS/United States
- P01 AR049084-01/AR/NIAMS NIH HHS/United States
- K08 AI083790-01/AI/NIAID NIH HHS/United States
- RC1 AR058621/AR/NIAMS NIH HHS/United States
- R01 AI070983/AI/NIAID NIH HHS/United States
- AI071651/AI/NIAID NIH HHS/United States
- P30 AR048311/AR/NIAMS NIH HHS/United States
- R01 AI063274-02/AI/NIAID NIH HHS/United States
- R01 AR043814/AR/NIAMS NIH HHS/United States
- R37 AI024717/AI/NIAID NIH HHS/United States
- R01 AR042460/AR/NIAMS NIH HHS/United States
- R01 AR033062/AR/NIAMS NIH HHS/United States
- R01CA141700/CA/NCI NIH HHS/United States
- P01 AI083194-01/AI/NIAID NIH HHS/United States
- UL1 RR025014/RR/NCRR NIH HHS/United States
- L30 AI071651/AI/NIAID NIH HHS/United States
- R01 AR43814/AR/NIAMS NIH HHS/United States
- R01 AR043727/AR/NIAMS NIH HHS/United States
- ARC_/Arthritis Research UK/United Kingdom
- R01 CA141700/CA/NCI NIH HHS/United States
- R37 24717/PHS HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R37 AI024717-17A1/AI/NIAID NIH HHS/United States
- P30 AR048311-07/AR/NIAMS NIH HHS/United States
- UL1 RR025014-02/RR/NCRR NIH HHS/United States
- R01 AR33062/AR/NIAMS NIH HHS/United States
- R21 AI070304/AI/NIAID NIH HHS/United States
- P01 AI083194/AI/NIAID NIH HHS/United States
- L30 AI071651-01/AI/NIAID NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- ULI RR025014-02/RR/NCRR NIH HHS/United States
- R01 AR051545/AR/NIAMS NIH HHS/United States
- UL1 RR024999-01/RR/NCRR NIH HHS/United States
- R01 AR043274/AR/NIAMS NIH HHS/United States
- N01 AI050026/AI/NIAID NIH HHS/United States
- R01 CA141700-01/CA/NCI NIH HHS/United States
- R01 AR051545-01A2/AR/NIAMS NIH HHS/United States
- N01 AR62277/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

