Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy
- PMID: 21333526
- PMCID: PMC3088881
- DOI: 10.1016/j.sbi.2011.01.008
Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy
Abstract
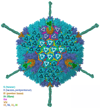
Nine different near-atomic resolution structures of icosahedral viruses, determined by electron cryo-microscopy and published between early 2008 and late 2010, fulfil predictions made 15 years ago that single-particle cryo-EM techniques could visualize molecular detail at 3-4Å resolution. This review summarizes technical developments, both in instrumentation and in computation, that have led to the new structures, which advance our understanding of virus assembly and cell entry.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
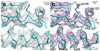
Similar articles
-
Single particle cryo-electron microscopy and 3-D reconstruction of viruses.Methods Mol Biol. 2014;1117:401-43. doi: 10.1007/978-1-62703-776-1_19. Methods Mol Biol. 2014. PMID: 24357374 Free PMC article.
-
Advances in computational approaches to structure determination of alphaviruses and flaviviruses using cryo-electron microscopy.J Struct Biol. 2023 Sep;215(3):107993. doi: 10.1016/j.jsb.2023.107993. Epub 2023 Jul 4. J Struct Biol. 2023. PMID: 37414374 Review.
-
Atomic cryo-EM structures of viruses.Curr Opin Struct Biol. 2017 Oct;46:122-129. doi: 10.1016/j.sbi.2017.07.002. Epub 2017 Aug 5. Curr Opin Struct Biol. 2017. PMID: 28787658 Free PMC article. Review.
-
Near-atomic-resolution cryo-EM for molecular virology.Curr Opin Virol. 2011 Aug;1(2):110-7. doi: 10.1016/j.coviro.2011.05.019. Curr Opin Virol. 2011. PMID: 21845206 Free PMC article. Review.
-
Cryogenic electron microscopy and single-particle analysis.Annu Rev Biochem. 2015;84:499-517. doi: 10.1146/annurev-biochem-060614-034226. Epub 2015 Feb 26. Annu Rev Biochem. 2015. PMID: 25747402 Review.
Cited by
-
Protein secondary structure determination by constrained single-particle cryo-electron tomography.Structure. 2012 Dec 5;20(12):2003-13. doi: 10.1016/j.str.2012.10.016. Structure. 2012. PMID: 23217682 Free PMC article.
-
Popping the cork: mechanisms of phage genome ejection.Nat Rev Microbiol. 2013 Mar;11(3):194-204. doi: 10.1038/nrmicro2988. Epub 2013 Feb 4. Nat Rev Microbiol. 2013. PMID: 23385786 Review.
-
Fabrication of Monolayer Graphene-Coated Grids for Cryoelectron Microscopy.J Vis Exp. 2023 Sep 8;(199):10.3791/65702. doi: 10.3791/65702. J Vis Exp. 2023. PMID: 37747197 Free PMC article.
-
Single particle analysis integrated with microscopy: a high-throughput approach for reconstructing icosahedral particles.J Struct Biol. 2014 Apr;186(1):8-18. doi: 10.1016/j.jsb.2014.02.016. Epub 2014 Mar 5. J Struct Biol. 2014. PMID: 24613762 Free PMC article.
-
Mixed-state electron ptychography enables sub-angstrom resolution imaging with picometer precision at low dose.Nat Commun. 2020 Jun 12;11(1):2994. doi: 10.1038/s41467-020-16688-6. Nat Commun. 2020. PMID: 32533001 Free PMC article.
References
-
-
Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. Reports a 3.8 Å resolution map of the rotavirus double-layered particle, validated in molecular detail by comparison with a crystal structure of the same particle.
-
-
-
Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. Reports a 4.5 Å resolution map of bacteriophage ε15. Two proteins make up a heterodimeric protomer in the T=7 icosahedral surface lattice. The secondary structure and approximate chain trace of one of the two proteins establishes its homology with the coat protein of bacteriophage HK97.
-
-
-
Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. Reports a 3.9 Å resolution map of cytoplasmic polyhedrosis virus, with a
de novo polypeptide chain tracing of the component proteins.
-
-
-
Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. Reports a structure at 4.2 Å resolution of a VP7-recoated, rotavirus double-layered particle (DLP) and shows how trimers of VP7, one of the two viral outer-layer proteins, fit onto trimers of VP6, the DLP structural component arrayed in a T=13 l icosahedral lattice.
-
-
-
Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. A 3.6 Å resolution map of a bovine papillomavirus particle yields a complete atomic model of the 72-pentamer icosahedral shell, allowing refinement of coordinates (R=37.7%, 15-3.6 Å). The structure corrects and extends an earlier model for key interactions in papillomavirus particle assembly.
-
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

