Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response
- PMID: 21325420
- PMCID: PMC3126222
- DOI: 10.1128/JVI.02232-10
Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response
Abstract
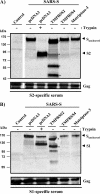
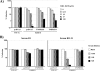
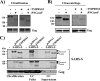
The spike (S) protein of the severe acute respiratory syndrome coronavirus (SARS-CoV) can be proteolytically activated by cathepsins B and L upon viral uptake into target cell endosomes. In contrast, it is largely unknown whether host cell proteases located in the secretory pathway of infected cells and/or on the surface of target cells can cleave SARS S. We along with others could previously show that the type II transmembrane protease TMPRSS2 activates the influenza virus hemagglutinin and the human metapneumovirus F protein by cleavage. Here, we assessed whether SARS S is proteolytically processed by TMPRSS2. Western blot analysis revealed that SARS S was cleaved into several fragments upon coexpression of TMPRSS2 (cis-cleavage) and upon contact between SARS S-expressing cells and TMPRSS2-positive cells (trans-cleavage). cis-cleavage resulted in release of SARS S fragments into the cellular supernatant and in inhibition of antibody-mediated neutralization, most likely because SARS S fragments function as antibody decoys. trans-cleavage activated SARS S on effector cells for fusion with target cells and allowed efficient SARS S-driven viral entry into targets treated with a lysosomotropic agent or a cathepsin inhibitor. Finally, ACE2, the cellular receptor for SARS-CoV, and TMPRSS2 were found to be coexpressed by type II pneumocytes, which represent important viral target cells, suggesting that SARS S is cleaved by TMPRSS2 in the lung of SARS-CoV-infected individuals. In summary, we show that TMPRSS2 might promote viral spread and pathogenesis by diminishing viral recognition by neutralizing antibodies and by activating SARS S for cell-cell and virus-cell fusion.
Figures

Similar articles
-
Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease.J Virol. 2011 Dec;85(24):13363-72. doi: 10.1128/JVI.05300-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994442 Free PMC article.
-
Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2.J Virol. 2010 Dec;84(24):12658-64. doi: 10.1128/JVI.01542-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926566 Free PMC article.
-
TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein.J Virol. 2014 Jan;88(2):1293-307. doi: 10.1128/JVI.02202-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227843 Free PMC article.
-
[Protease-dependent cell entry mechanism of coronaviruses].Uirusu. 2011 Jun;61(1):109-16. doi: 10.2222/jsv.61.109. Uirusu. 2011. PMID: 21972562 Review. Japanese.
-
Molecular mechanisms for understanding the association between TMPRSS2 and beta coronaviruses SARS-CoV-2, SARS-CoV and MERS-CoV infection: scoping review.Arch Microbiol. 2021 Dec 25;204(1):77. doi: 10.1007/s00203-021-02727-3. Arch Microbiol. 2021. PMID: 34953136 Free PMC article. Review.
Cited by
-
Aromatase, testosterone, TMPRSS2: determinants of COVID-19 severity.Biol Sex Differ. 2024 Oct 24;15(1):84. doi: 10.1186/s13293-024-00658-4. Biol Sex Differ. 2024. PMID: 39449074 Free PMC article.
-
Potential Effects of Hyperglycemia on SARS-CoV-2 Entry Mechanisms in Pancreatic Beta Cells.Viruses. 2024 Aug 2;16(8):1243. doi: 10.3390/v16081243. Viruses. 2024. PMID: 39205219 Free PMC article. Review.
-
The Mechanism of Anti-Viral Activity of a Novel, Hydroponically Selenium-Enriched Garlic Powder (SelenoForce®) Against SARS-CoV-2 Virus.Glob Adv Integr Med Health. 2024 Aug 8;13:27536130241268100. doi: 10.1177/27536130241268100. eCollection 2024 Jan-Dec. Glob Adv Integr Med Health. 2024. PMID: 39130207 Free PMC article.
-
Feline coronavirus influences the biogenesis and composition of extracellular vesicles derived from CRFK cells.Front Vet Sci. 2024 Jul 18;11:1388438. doi: 10.3389/fvets.2024.1388438. eCollection 2024. Front Vet Sci. 2024. PMID: 39091390 Free PMC article.
-
Potential Protective Factors for Allergic Rhinitis Patients Infected with COVID-19.Curr Issues Mol Biol. 2024 Jun 28;46(7):6633-6645. doi: 10.3390/cimb46070395. Curr Issues Mol Biol. 2024. PMID: 39057037 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

