A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation
- PMID: 21324897
- PMCID: PMC3069436
- DOI: 10.1074/jbc.M110.176099
A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation
Abstract
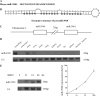
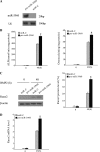
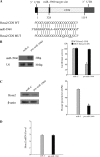
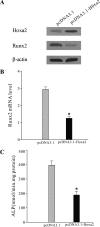
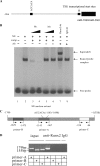
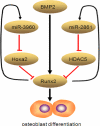
Our recent study showed that miR-2861 promotes osteoblast differentiation by targeting histone deacetylase 5, resulting in increased runt-related transcription factor 2 (Runx2) protein production. Here we identified another new microRNA (miRNA) (miR-3960) that played a regulatory role in osteoblast differentiation through a regulatory feedback loop with miR-2861. miR-3960 and miR-2861 were found clustered at the same loci. miR-3960 was transcribed during bone morphogenic protein 2 (BMP2)-induced osteogenesis of ST2 stromal cells. Overexpression of miR-3960 promoted BMP2-induced osteoblastogenesis. However, the inhibition of miR-3960 expression attenuated the osteoblastogenesis. Homeobox A2 (Hoxa2), a repressor of Runx2 expression, was confirmed to be a target of miR-3960. Electrophoretic mobility shift assay and chromatin immunoprecipitation experiments confirmed that Runx2 bound to the promoter of the miR-3960/miR-2861 cluster. Furthermore, overexpression of Runx2 induced miR-3960/miR-2861 transcription, and block of Runx2 expression attenuated BMP2-induced miR-3960/miR-2861 transcription. Here we report that miR-3960 and miR-2861, transcribed together from the same miRNA polycistron, both function in osteoblast differentiation through a novel Runx2/miR-3960/miR-2861 regulatory feedback loop. Our findings provide new insights into the roles of miRNAs in osteoblast differentiation.
Figures


Similar articles
-
A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans.J Clin Invest. 2009 Dec;119(12):3666-77. doi: 10.1172/JCI39832. Epub 2009 Nov 16. J Clin Invest. 2009. PMID: 19920351 Free PMC article.
-
A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program.Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19879-84. doi: 10.1073/pnas.1007698107. Epub 2010 Oct 27. Proc Natl Acad Sci U S A. 2010. PMID: 20980664 Free PMC article.
-
Runx2/miR-3960/miR-2861 Positive Feedback Loop Is Responsible for Osteogenic Transdifferentiation of Vascular Smooth Muscle Cells.Biomed Res Int. 2015;2015:624037. doi: 10.1155/2015/624037. Epub 2015 Jun 28. Biomed Res Int. 2015. PMID: 26221600 Free PMC article.
-
Regulation of Runx2 by MicroRNAs in osteoblast differentiation.Life Sci. 2019 Sep 1;232:116676. doi: 10.1016/j.lfs.2019.116676. Epub 2019 Jul 21. Life Sci. 2019. PMID: 31340165 Review.
-
Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development.Int J Mol Sci. 2023 Feb 3;24(3):2979. doi: 10.3390/ijms24032979. Int J Mol Sci. 2023. PMID: 36769300 Free PMC article. Review.
Cited by
-
MiR-17-3p inhibits osteoblast differentiation by downregulating Sox6 expression.FEBS Open Bio. 2020 Nov;10(11):2499-2506. doi: 10.1002/2211-5463.12979. Epub 2020 Oct 25. FEBS Open Bio. 2020. PMID: 32946669 Free PMC article.
-
Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix.PLoS One. 2013;8(3):e58104. doi: 10.1371/journal.pone.0058104. Epub 2013 Mar 5. PLoS One. 2013. PMID: 23472141 Free PMC article.
-
MicroRNA functions in osteogenesis and dysfunctions in osteoporosis.Curr Osteoporos Rep. 2013 Jun;11(2):72-82. doi: 10.1007/s11914-013-0143-6. Curr Osteoporos Rep. 2013. PMID: 23605904 Free PMC article. Review.
-
A microRNA Approach to Discriminate Cortical Low Bone Turnover in Renal Osteodystrophy.JBMR Plus. 2020 Mar 25;4(5):e10353. doi: 10.1002/jbm4.10353. eCollection 2020 May. JBMR Plus. 2020. PMID: 32490328 Free PMC article.
-
Associations of polymorphisms in the SOST gene and bone mineral density in postmenopausal Chinese Women.Osteoporos Int. 2014 Dec;25(12):2797-803. doi: 10.1007/s00198-014-2832-0. Epub 2014 Aug 8. Osteoporos Int. 2014. PMID: 25103216
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

