Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1
- PMID: 21300889
- PMCID: PMC3044381
- DOI: 10.1073/pnas.1008098108
Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1
Abstract
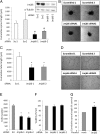
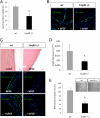
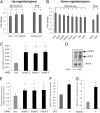
JmjC domain-containing proteins play a crucial role in the control of gene expression by acting as protein hydroxylases or demethylases, thereby controlling histone methylation or splicing. Here, we demonstrate that silencing of Jumonji domain-containing protein 6 (Jmjd6) impairs angiogenic functions of endothelial cells by changing the gene expression and modulating the splicing of the VEGF-receptor 1 (Flt1). Reduction of Jmjd6 expression altered splicing of Flt1 and increased the levels of the soluble form of Flt1, which binds to VEGF and placental growth factor (PlGF) and thereby inhibits angiogenesis. Saturating VEGF or PlGF or neutralizing antibodies directed against soluble Flt1 rescued the angiogenic defects induced by Jmjd6 silencing. Jmjd6 interacts with the splicing factors U2AF65 that binds to Flt1 mRNA. In conclusion, Jmjd6 regulates the splicing of Flt1, thereby controlling angiogenic sprouting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
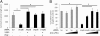
Similar articles
-
Jumonji Domain Containing Protein 6 Is Decreased in Human Preeclamptic Placentas and Regulates sFLT-1 Splice Variant Production.Biol Reprod. 2016 Mar;94(3):59. doi: 10.1095/biolreprod.115.134460. Epub 2016 Jan 27. Biol Reprod. 2016. PMID: 26819475
-
Differential regulation of sFlt-1 splicing by U2AF65 and JMJD6 in placental-derived and endothelial cells.Biosci Rep. 2020 Feb 28;40(2):BSR20193252. doi: 10.1042/BSR20193252. Biosci Rep. 2020. PMID: 32039444 Free PMC article.
-
Role of PlGF in the intra- and intermolecular cross talk between the VEGF receptors Flt1 and Flk1.Nat Med. 2003 Jul;9(7):936-43. doi: 10.1038/nm884. Nat Med. 2003. PMID: 12796773
-
Insights into Jumonji C-domain containing protein 6 (JMJD6): a multifactorial role in foot-and-mouth disease virus replication in cells.Virus Genes. 2017 Jun;53(3):340-351. doi: 10.1007/s11262-017-1449-8. Epub 2017 Mar 31. Virus Genes. 2017. PMID: 28364140 Review.
-
Regulatory role of JMJD6 in placental development.Expert Rev Mol Med. 2022 Sep 19;24:e34. doi: 10.1017/erm.2022.30. Expert Rev Mol Med. 2022. PMID: 36222080 Review.
Cited by
-
Protein Hydroxylation Catalyzed by 2-Oxoglutarate-dependent Oxygenases.J Biol Chem. 2015 Aug 21;290(34):20712-20722. doi: 10.1074/jbc.R115.662627. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152730 Free PMC article. Review.
-
Role of Arginine Methylation in Alternative Polyadenylation of VEGFR-1 (Flt-1) pre-mRNA.Int J Mol Sci. 2020 Sep 4;21(18):6460. doi: 10.3390/ijms21186460. Int J Mol Sci. 2020. PMID: 32899690 Free PMC article. Review.
-
Genetic Regulation of Neuronal Progranulin Reveals a Critical Role for the Autophagy-Lysosome Pathway.J Neurosci. 2019 Apr 24;39(17):3332-3344. doi: 10.1523/JNEUROSCI.3498-17.2019. Epub 2019 Jan 29. J Neurosci. 2019. PMID: 30696728 Free PMC article.
-
The unfolded protein response in the protozoan parasite Toxoplasma gondii features translational and transcriptional control.Eukaryot Cell. 2013 Jul;12(7):979-89. doi: 10.1128/EC.00021-13. Epub 2013 May 10. Eukaryot Cell. 2013. PMID: 23666622 Free PMC article.
-
PCAF-mediated acetylation of transcriptional factor HOXB9 suppresses lung adenocarcinoma progression by targeting oncogenic protein JMJD6.Nucleic Acids Res. 2016 Dec 15;44(22):10662-10675. doi: 10.1093/nar/gkw808. Epub 2016 Sep 8. Nucleic Acids Res. 2016. PMID: 27613418 Free PMC article.
References
-
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. - PubMed
-
- Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. - PubMed
-
- Hewitson KS, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. - PubMed
-
- Pollard PJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

