Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1
- PMID: 21300781
- PMCID: PMC3126331
- DOI: 10.1128/MCB.01284-10
Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1
Abstract
In budding yeast and humans, cohesion establishment during S phase requires the acetyltransferase Eco1/Esco1-2, which acetylates the cohesin subunit Smc3 on two conserved lysine residues. Whether Smc3 is the sole Eco1/Esco1-2 effector and how Smc3 acetylation promotes cohesion are unknown. In fission yeast (Schizosaccharomyces pombe), as in humans, cohesin binding to G(1) chromosomes is dynamic and the unloading reaction is stimulated by Wpl1 (human ortholog, Wapl). During S phase, a subpopulation of cohesin becomes stably bound to chromatin in an Eso1 (fission yeast Eco1/Esco1-2)-dependent manner. Cohesin stabilization occurs unevenly along chromosomes. Cohesin remains largely labile at the rDNA repeats but binds mostly in the stable mode to pericentromere regions. This pattern is largely unchanged in eso1Δ wpl1Δ cells, and cohesion is unaffected, indicating that the main Eso1 role is counteracting Wpl1. A mutant of Psm3 (fission yeast Smc3) that mimics its acetylated state renders cohesin less sensitive to Wpl1-dependent unloading and partially bypasses the Eso1 requirement but cannot generate the stable mode of cohesin binding in the absence of Eso1. Conversely, nonacetylatable Psm3 reduces the stable cohesin fraction and affects cohesion in a Wpl1-dependent manner, but cells are viable. We propose that Psm3 acetylation contributes to Eso1 counteracting of Wpl1 to secure stable cohesin interaction with postreplicative chromosomes but that it is not the sole molecular event by which this occurs.
Figures






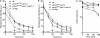
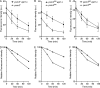
Similar articles
-
Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction.EMBO Rep. 2012 Jun 29;13(7):645-52. doi: 10.1038/embor.2012.72. EMBO Rep. 2012. PMID: 22640989 Free PMC article.
-
A second Wpl1 anti-cohesion pathway requires dephosphorylation of fission yeast kleisin Rad21 by PP4.EMBO J. 2017 May 15;36(10):1364-1378. doi: 10.15252/embj.201696050. Epub 2017 Apr 24. EMBO J. 2017. PMID: 28438891 Free PMC article.
-
The acetyltransferase Eco1 elicits cohesin dimerization during S phase.J Biol Chem. 2020 May 29;295(22):7554-7565. doi: 10.1074/jbc.RA120.013102. Epub 2020 Apr 20. J Biol Chem. 2020. PMID: 32312753 Free PMC article.
-
Cohesin codes - interpreting chromatin architecture and the many facets of cohesin function.J Cell Sci. 2013 Jan 1;126(Pt 1):31-41. doi: 10.1242/jcs.116566. J Cell Sci. 2013. PMID: 23516328 Free PMC article. Review.
-
The expanding phenotypes of cohesinopathies: one ring to rule them all!Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
-
The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion.EMBO J. 2018 Aug 1;37(15):e97150. doi: 10.15252/embj.201797150. Epub 2018 Jun 21. EMBO J. 2018. PMID: 29930102 Free PMC article.
-
Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast.EMBO Rep. 2011 Oct 28;12(11):1189-95. doi: 10.1038/embor.2011.188. EMBO Rep. 2011. PMID: 21979813 Free PMC article.
-
Pds5A and Pds5B Display Non-redundant Functions in Mitosis and Their Loss Triggers Chk1 Activation.Front Cell Dev Biol. 2020 Jul 14;8:531. doi: 10.3389/fcell.2020.00531. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32760717 Free PMC article.
-
Sister chromatid cohesion.Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a011130. doi: 10.1101/cshperspect.a011130. Cold Spring Harb Perspect Biol. 2012. PMID: 23043155 Free PMC article. Review.
-
Whole-Genome Sequencing of Suppressor DNA Mixtures Identifies Pathways That Compensate for Chromosome Segregation Defects in Schizosaccharomyces pombe.G3 (Bethesda). 2018 Mar 2;8(3):1031-1038. doi: 10.1534/g3.118.200048. G3 (Bethesda). 2018. PMID: 29352077 Free PMC article.
References
-
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218–233 - PubMed
-
- Arumugam P., et al. 2003. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 13:1941–1953 - PubMed
-
- Arumugam P., Nishino T., Haering C. H., Gruber S., Nasmyth K. 2006. Cohesin's ATPase activity is stimulated by the C-terminal Winged-Helix domain of its kleisin subunit. Curr. Biol. 16:1998–2008 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
