Long tract of untranslated CAG repeats is deleterious in transgenic mice
- PMID: 21283659
- PMCID: PMC3025035
- DOI: 10.1371/journal.pone.0016417
Long tract of untranslated CAG repeats is deleterious in transgenic mice
Abstract
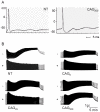
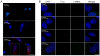
The most frequent trinucleotide repeat found in human disorders is the CAG sequence. Expansion of CAG repeats is mostly found in coding regions and is thought to cause diseases through a protein mechanism. Recently, expanded CAG repeats were shown to induce toxicity at the RNA level in Drosophila and C. elegans. These findings raise the possibility that CAG repeats may trigger RNA-mediated pathogenesis in mammals. Here, we demonstrate that transgenic mice expressing EGFP transcripts with long CAG repeats in the 3' untranslated region develop pathogenic features. Expression of the transgene was directed to the muscle in order to compare the resulting phenotype to that caused by the CUG expansion, as occurs in myotonic dystrophy. Transgenic mice expressing 200, but not those expressing 0 or 23 CAG repeats, showed alterations in muscle morphology, histochemistry and electrophysiology, as well as abnormal behavioral phenotypes. Expression of the expanded CAG repeats in testes resulted in reduced fertility due to defective sperm motility. The production of EGFP protein was significantly reduced by the 200 CAG repeats, and no polyglutamine-containing product was detected, which argues against a protein mechanism. Moreover, nuclear RNA foci were detected for the long CAG repeats. These data support the notion that expanded CAG repeat RNA can cause deleterious effects in mammals. They also suggest the possible involvement of an RNA mechanism in human diseases with long CAG repeats.
Conflict of interest statement
Figures
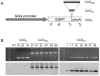

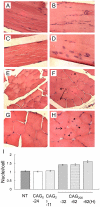

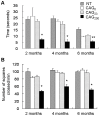
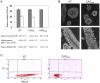
Similar articles
-
CAG repeats mimic CUG repeats in the misregulation of alternative splicing.Nucleic Acids Res. 2011 Nov 1;39(20):8938-51. doi: 10.1093/nar/gkr608. Epub 2011 Jul 27. Nucleic Acids Res. 2011. PMID: 21795378 Free PMC article.
-
Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat.Science. 2000 Sep 8;289(5485):1769-73. doi: 10.1126/science.289.5485.1769. Science. 2000. PMID: 10976074
-
Expanded CUG Repeats Trigger Disease Phenotype and Expression Changes through the RNAi Machinery in C. elegans.J Mol Biol. 2019 Apr 19;431(9):1711-1728. doi: 10.1016/j.jmb.2019.03.003. Epub 2019 Mar 14. J Mol Biol. 2019. PMID: 30878478 Review.
-
Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy.PLoS Genet. 2011 Mar;7(3):e1001340. doi: 10.1371/journal.pgen.1001340. Epub 2011 Mar 17. PLoS Genet. 2011. PMID: 21437269 Free PMC article.
-
CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders.RNA Biol. 2011 Jul-Aug;8(4):565-71. doi: 10.4161/rna.8.4.15397. Epub 2011 Jul 1. RNA Biol. 2011. PMID: 21593608 Free PMC article. Review.
Cited by
-
RNA-mediated pathogenic mechanisms in polyglutamine diseases and amyotrophic lateral sclerosis.Front Cell Neurosci. 2014 Dec 19;8:431. doi: 10.3389/fncel.2014.00431. eCollection 2014. Front Cell Neurosci. 2014. PMID: 25565965 Free PMC article. Review.
-
RNA-binding protein misregulation in microsatellite expansion disorders.Adv Exp Med Biol. 2014;825:353-88. doi: 10.1007/978-1-4939-1221-6_10. Adv Exp Med Biol. 2014. PMID: 25201111 Free PMC article. Review.
-
Small non-coding RNAs add complexity to the RNA pathogenic mechanisms in trinucleotide repeat expansion diseases.Front Mol Neurosci. 2013 Dec 3;6:45. doi: 10.3389/fnmol.2013.00045. Front Mol Neurosci. 2013. PMID: 24348326 Free PMC article. Review.
-
Small interfering RNAs based on huntingtin trinucleotide repeats are highly toxic to cancer cells.EMBO Rep. 2018 Mar;19(3):e45336. doi: 10.15252/embr.201745336. Epub 2018 Feb 12. EMBO Rep. 2018. PMID: 29440125 Free PMC article.
-
Current Status of Gene Therapy Research in Polyglutamine Spinocerebellar Ataxias.Int J Mol Sci. 2021 Apr 19;22(8):4249. doi: 10.3390/ijms22084249. Int J Mol Sci. 2021. PMID: 33921915 Free PMC article. Review.
References
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
-
- Everett CM, Wood NW. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385–2405. - PubMed
-
- Ranum LP, Day JW. Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet. 2004;20:506–512. - PubMed
-
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. - PubMed
-
- Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15 Spec No 2:R162–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

