Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4
- PMID: 21270136
- PMCID: PMC3062185
- DOI: 10.1261/rna.2455411
Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4
Abstract
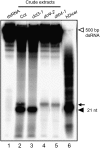
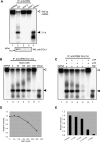
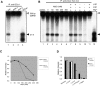
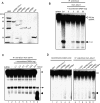
Arabidopsis thaliana Dicer-like 4 (DCL4) produces 21-nt small interfering RNAs from both endogenous and exogenous double-stranded RNAs (dsRNAs), and it interacts with DRB4, a dsRNA-binding protein, in vivo and in vitro. However, the role of DRB4 in DCL4 activity remains unclear because the dsRNA-cleaving activity of DCL4 has not been characterized biochemically. In this study, we biochemically characterize DCL4's Dicer activity and establish that DRB4 is required for this activity in vitro. Crude extracts from Arabidopsis seedlings cleave long dsRNAs into 21-nt small RNAs in a DCL4/DRB4-dependent manner. Immunoaffinity-purified DCL4 complexes produce 21-nt small RNAs from long dsRNA, and these complexes have biochemical properties similar to those of known Dicer family proteins. The DCL4 complexes purified from drb4-1 do not cleave dsRNA, and the addition of recombinant DRB4 to drb4-1 complexes specifically recovers the 21-nt small RNA generation. These results reveal that DCL4 requires DRB4 to cleave long dsRNA into 21-nt small RNAs in vitro. Amino acid substitutions in conserved dsRNA-binding domains (dsRBDs) of DRB4 impair three activities: binding to dsRNA, interacting with DCL4, and facilitating DCL4 activity. These observations indicate that the dsRBDs are critical for DRB4 function. Our biochemical approach and observations clearly show that DRB4 is specifically required for DCL4 activity in vitro.
Figures
Similar articles
-
Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana.Plant Mol Biol. 2005 Jan;57(2):173-88. doi: 10.1007/s11103-004-6853-5. Plant Mol Biol. 2005. PMID: 15821876
-
The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway.Plant Mol Biol. 2007 Apr;63(6):777-85. doi: 10.1007/s11103-006-9125-8. Epub 2007 Jan 14. Plant Mol Biol. 2007. PMID: 17221360
-
A complex of Arabidopsis DRB proteins can impair dsRNA processing.RNA. 2017 May;23(5):782-797. doi: 10.1261/rna.059519.116. Epub 2017 Feb 23. RNA. 2017. PMID: 28232389 Free PMC article.
-
Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis.J Plant Res. 2017 Jan;130(1):33-44. doi: 10.1007/s10265-016-0877-1. Epub 2016 Nov 24. J Plant Res. 2017. PMID: 27885504 Review.
-
The mechanism of RNase III action: how dicer dices.Curr Top Microbiol Immunol. 2008;320:99-116. doi: 10.1007/978-3-540-75157-1_5. Curr Top Microbiol Immunol. 2008. PMID: 18268841 Review.
Cited by
-
Parallel action of AtDRB2 and RdDM in the control of transposable element expression.BMC Plant Biol. 2015 Mar 3;15:70. doi: 10.1186/s12870-015-0455-z. BMC Plant Biol. 2015. PMID: 25849103 Free PMC article.
-
Double-Stranded-RNA-Binding Protein 2 Participates in Antiviral Defense.J Virol. 2020 May 18;94(11):e00017-20. doi: 10.1128/JVI.00017-20. Print 2020 May 18. J Virol. 2020. PMID: 32213615 Free PMC article.
-
'Black sheep' that don't leave the double-stranded RNA-binding domain fold.Trends Biochem Sci. 2014 Jul;39(7):328-40. doi: 10.1016/j.tibs.2014.05.003. Epub 2014 Jun 19. Trends Biochem Sci. 2014. PMID: 24954387 Free PMC article. Review.
-
APC/C-mediated degradation of dsRNA-binding protein 4 (DRB4) involved in RNA silencing.PLoS One. 2012;7(4):e35173. doi: 10.1371/journal.pone.0035173. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545099 Free PMC article.
-
Suppress to Survive-Implication of Plant Viruses in PTGS.Plant Mol Biol Report. 2015;33(3):335-346. doi: 10.1007/s11105-014-0755-8. Plant Mol Biol Report. 2015. PMID: 25999662 Free PMC article. Review.
References
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 - PubMed
-
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett 582: 2753–2760 - PubMed
-
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O 2006. Hierarchical action and inhibition of plant DICER-LIKE proteins in antiviral defence. Science 313: 68–71 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
