Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs
- PMID: 21258325
- PMCID: PMC3075553
- DOI: 10.1038/nsmb.1975
Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs
Erratum in
- Nat Struct Mol Biol. 2012 Mar;19(3):364
Abstract
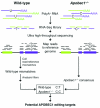
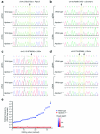
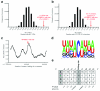
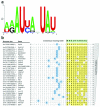
Apolipoprotein B-editing enzyme, catalytic polypeptide-1 (APOBEC1) is a cytidine deaminase initially identified by its activity in converting a specific cytidine (C) to uridine (U) in apolipoprotein B (apoB) mRNA transcripts in the small intestine. Editing results in the translation of a truncated apoB isoform with distinct functions in lipid transport. To address the possibility that APOBEC1 edits additional mRNAs, we developed a transcriptome-wide comparative RNA sequencing (RNA-Seq) screen. We identified and validated 32 previously undescribed mRNA targets of APOBEC1 editing, all of which are located in AU-rich segments of transcript 3' untranslated regions (3' UTRs). Further analysis established several characteristic sequence features of editing targets, which were predictive for the identification of additional APOBEC1 substrates. The transcriptomics approach to RNA editing presented here dramatically expands the list of APOBEC1 mRNA editing targets and reveals a novel cellular mechanism for the modification of transcript 3' UTRs.
Figures
Similar articles
-
Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice. Structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases.J Biol Chem. 1995 Jun 2;270(22):13042-56. doi: 10.1074/jbc.270.22.13042. J Biol Chem. 1995. PMID: 7768898
-
Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver.Genome Biol. 2014 Jun 19;15(6):R79. doi: 10.1186/gb-2014-15-6-r79. Genome Biol. 2014. PMID: 24946870 Free PMC article.
-
Intestine-specific expression of Apobec-1 rescues apolipoprotein B RNA editing and alters chylomicron production in Apobec1 -/- mice.J Lipid Res. 2012 Dec;53(12):2643-55. doi: 10.1194/jlr.M030494. Epub 2012 Sep 19. J Lipid Res. 2012. PMID: 22993231 Free PMC article.
-
Apobec-1 and apolipoprotein B mRNA editing.Biochim Biophys Acta. 1997 Mar 10;1345(1):11-26. doi: 10.1016/s0005-2760(96)00156-7. Biochim Biophys Acta. 1997. PMID: 9084497 Review.
-
RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA.Biochim Biophys Acta. 2000 Nov 15;1494(1-2):1-13. doi: 10.1016/s0167-4781(00)00219-0. Biochim Biophys Acta. 2000. PMID: 11072063 Review.
Cited by
-
Global RNA editome landscape discovers reduced RNA editing in glioma: loss of editing of gamma-amino butyric acid receptor alpha subunit 3 (GABRA3) favors glioma migration and invasion.PeerJ. 2020 Sep 29;8:e9755. doi: 10.7717/peerj.9755. eCollection 2020. PeerJ. 2020. PMID: 33062411 Free PMC article.
-
AID/APOBEC-network reconstruction identifies pathways associated with survival in ovarian cancer.BMC Genomics. 2016 Aug 16;17(1):643. doi: 10.1186/s12864-016-3001-y. BMC Genomics. 2016. PMID: 27527602 Free PMC article.
-
The Roles of APOBEC-mediated RNA Editing in SARS-CoV-2 Mutations, Replication and Fitness.bioRxiv [Preprint]. 2022 Apr 7:2021.12.18.473309. doi: 10.1101/2021.12.18.473309. bioRxiv. 2022. Update in: Sci Rep. 2022 Sep 13;12(1):14972. doi: 10.1038/s41598-022-19067-x. PMID: 34981048 Free PMC article. Updated. Preprint.
-
A tour through the transcriptional landscape of platelets.Blood. 2014 Jul 24;124(4):493-502. doi: 10.1182/blood-2014-04-512756. Epub 2014 Jun 5. Blood. 2014. PMID: 24904119 Free PMC article. Review.
-
Effective tools for RNA-derived therapeutics: siRNA interference or miRNA mimicry.Theranostics. 2021 Aug 11;11(18):8771-8796. doi: 10.7150/thno.62642. eCollection 2021. Theranostics. 2021. PMID: 34522211 Free PMC article. Review.
References
-
- Grosjean H, et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. - PubMed
-
- Gray MW. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life. 2003;55:227–33. - PubMed
-
- Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA-editing. RNA biology. 2010;7 - PubMed
-
- Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

