Nascent transcript sequencing visualizes transcription at nucleotide resolution
- PMID: 21248844
- PMCID: PMC3880149
- DOI: 10.1038/nature09652
Nascent transcript sequencing visualizes transcription at nucleotide resolution
Abstract
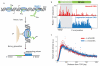
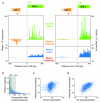
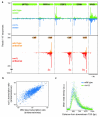
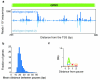
Recent studies of transcription have revealed a level of complexity not previously appreciated even a few years ago, both in the intricate use of post-initiation control and the mass production of rapidly degraded transcripts. Dissection of these pathways requires strategies for precisely following transcripts as they are being produced. Here we present an approach (native elongating transcript sequencing, NET-seq), based on deep sequencing of 3' ends of nascent transcripts associated with RNA polymerase, to monitor transcription at nucleotide resolution. Application of NET-seq in Saccharomyces cerevisiae reveals that although promoters are generally capable of divergent transcription, the Rpd3S deacetylation complex enforces strong directionality to most promoters by suppressing antisense transcript initiation. Our studies also reveal pervasive polymerase pausing and backtracking throughout the body of transcripts. Average pause density shows prominent peaks at each of the first four nucleosomes, with the peak location occurring in good agreement with in vitro biophysical measurements. Thus, nucleosome-induced pausing represents a major barrier to transcriptional elongation in vivo.
Figures
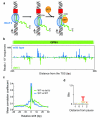
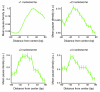
Comment in
-
Gene expression: Teasing out transcription.Nat Rev Mol Cell Biol. 2011 Mar;12(3):138-9. doi: 10.1038/nrm3066. Epub 2011 Feb 9. Nat Rev Mol Cell Biol. 2011. PMID: 21304551 No abstract available.
-
Gene expression: Teasing out transcription.Nat Rev Genet. 2011 Mar;12(3):152. doi: 10.1038/nrg2959. Nat Rev Genet. 2011. PMID: 21331084 No abstract available.
Similar articles
-
The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae.J Biol Chem. 2012 Aug 17;287(34):29021-34. doi: 10.1074/jbc.M112.380147. Epub 2012 Jul 2. J Biol Chem. 2012. PMID: 22761425 Free PMC article.
-
Widespread bidirectional promoters are the major source of cryptic transcripts in yeast.Nature. 2009 Feb 19;457(7232):1038-42. doi: 10.1038/nature07747. Epub 2009 Jan 25. Nature. 2009. PMID: 19169244
-
Bidirectional promoters generate pervasive transcription in yeast.Nature. 2009 Feb 19;457(7232):1033-7. doi: 10.1038/nature07728. Epub 2009 Jan 25. Nature. 2009. PMID: 19169243 Free PMC article.
-
Histone acetylation and deacetylation in yeast.Nat Rev Mol Cell Biol. 2003 Apr;4(4):276-84. doi: 10.1038/nrm1075. Nat Rev Mol Cell Biol. 2003. PMID: 12671650 Review.
-
The specificity of H2A.Z occupancy in the yeast genome and its relationship to transcription.Curr Genet. 2020 Oct;66(5):939-944. doi: 10.1007/s00294-020-01087-7. Epub 2020 Jun 14. Curr Genet. 2020. PMID: 32537667 Review.
Cited by
-
Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans.BMC Genomics. 2014 Dec 22;15(1):1157. doi: 10.1186/1471-2164-15-1157. BMC Genomics. 2014. PMID: 25534009 Free PMC article.
-
The RSC complex remodels nucleosomes in transcribed coding sequences and promotes transcription in Saccharomyces cerevisiae.Genetics. 2021 Apr 15;217(4):iyab021. doi: 10.1093/genetics/iyab021. Genetics. 2021. PMID: 33857307 Free PMC article.
-
Single-molecule studies of RNAPII elongation.Biochim Biophys Acta. 2013 Jan;1829(1):29-38. doi: 10.1016/j.bbagrm.2012.08.006. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982192 Free PMC article. Review.
-
Active chromatin and noncoding RNAs: an intimate relationship.Curr Opin Genet Dev. 2012 Apr;22(2):172-8. doi: 10.1016/j.gde.2011.11.002. Epub 2011 Dec 7. Curr Opin Genet Dev. 2012. PMID: 22154525 Free PMC article. Review.
-
The Determinants of Directionality in Transcriptional Initiation.Trends Genet. 2016 Jun;32(6):322-333. doi: 10.1016/j.tig.2016.03.005. Epub 2016 Apr 7. Trends Genet. 2016. PMID: 27066865 Free PMC article. Review.
References
-
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. - PubMed
-
- Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. - PubMed
-
- Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. - PubMed
-
- Rougvie AE, Lis JT. The RNA polymeraseII molecule at the end of the uninduced hsp70 gene of D.melanogaster is transcriptionallyengaged. Cell. 1988;54:795–804. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

