Control of protein function by reversible Nɛ-lysine acetylation in bacteria
- PMID: 21239213
- PMCID: PMC3078959
- DOI: 10.1016/j.mib.2010.12.013
Control of protein function by reversible Nɛ-lysine acetylation in bacteria
Abstract
Recently published work indicates that reversible N(ɛ)-lysine (N(ɛ)-Lys) acetylation of proteins in bacteria may be as diverse, and as important for cellular function, as it has been reported in eukaryotes for the last five decades. In addition to biochemical and genetic approaches, proteomic studies have identified N(ɛ)-Lys acetylation of proteins and enzymes involved in diverse cellular activities such as transcription, translation, stress response, detoxification, and especially carbohydrate and energy metabolism. These findings provide a platform for elucidating the molecular mechanisms behind modulation of enzyme activity by N(ɛ)-Lys acetylation, as well as for understanding how the prokaryotic cell maintains homeostasis in a changing environment.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Regulation of bacterial physiology by lysine acetylation of proteins.N Biotechnol. 2014 Dec 25;31(6):586-95. doi: 10.1016/j.nbt.2014.03.002. Epub 2014 Mar 15. N Biotechnol. 2014. PMID: 24636882 Review.
-
Protein Acetylation in Bacteria.Annu Rev Microbiol. 2019 Sep 8;73:111-132. doi: 10.1146/annurev-micro-020518-115526. Epub 2019 May 15. Annu Rev Microbiol. 2019. PMID: 31091420 Free PMC article. Review.
-
Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli.Mol Cell Proteomics. 2009 Feb;8(2):215-25. doi: 10.1074/mcp.M800187-MCP200. Epub 2008 Aug 23. Mol Cell Proteomics. 2009. PMID: 18723842 Free PMC article.
-
Genome-scale analysis of regulatory protein acetylation enzymes from photosynthetic eukaryotes.BMC Genomics. 2017 Jul 5;18(1):514. doi: 10.1186/s12864-017-3894-0. BMC Genomics. 2017. PMID: 28679357 Free PMC article.
-
Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis.Plant Physiol. 2011 Apr;155(4):1779-90. doi: 10.1104/pp.110.171595. Epub 2011 Feb 10. Plant Physiol. 2011. PMID: 21311031 Free PMC article.
Cited by
-
Glucose Induces ECF Sigma Factor Genes, sigX and sigM, Independent of Cognate Anti-sigma Factors through Acetylation of CshA in Bacillus subtilis.Front Microbiol. 2016 Nov 29;7:1918. doi: 10.3389/fmicb.2016.01918. eCollection 2016. Front Microbiol. 2016. PMID: 27965645 Free PMC article.
-
Post-translational Serine/Threonine Phosphorylation and Lysine Acetylation: A Novel Regulatory Aspect of the Global Nitrogen Response Regulator GlnR in S. coelicolor M145.Front Mol Biosci. 2016 Aug 9;3:38. doi: 10.3389/fmolb.2016.00038. eCollection 2016. Front Mol Biosci. 2016. PMID: 27556027 Free PMC article.
-
Studying the Lysine Acetylation of Malate Dehydrogenase.J Mol Biol. 2017 May 5;429(9):1396-1405. doi: 10.1016/j.jmb.2017.03.027. Epub 2017 Mar 31. J Mol Biol. 2017. PMID: 28366830 Free PMC article.
-
Modulation of Central Carbon Metabolism by Acetylation of Isocitrate Lyase in Mycobacterium tuberculosis.Sci Rep. 2017 Mar 21;7:44826. doi: 10.1038/srep44826. Sci Rep. 2017. PMID: 28322251 Free PMC article.
-
Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans.Mol Microbiol. 2013 Jan;87(1):152-67. doi: 10.1111/mmi.12088. Epub 2012 Nov 30. Mol Microbiol. 2013. PMID: 23199287 Free PMC article.
References
-
-
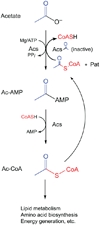
Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. The first report of sirtuin-dependent control of a metabolic enzyme.
-
-
- Brandl A, Heinzel T, Kramer OH. Histone deacetylases: salesmen and customers in the post-translational modification market. Biol. Cell. 2009;101:193–205. - PubMed
-
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 11:426–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

