The Biomolecular Interaction Network Database in PSI-MI 2.5
- PMID: 21233089
- PMCID: PMC3021793
- DOI: 10.1093/database/baq037
The Biomolecular Interaction Network Database in PSI-MI 2.5
Abstract
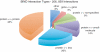
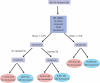
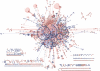
The Biomolecular Interaction Network Database (BIND) is a major source of curated biomolecular interactions, which has been unmaintained for the last few years, a trend which will eventually result in the loss of a significant amount of unique biomolecular interaction information, mostly as database identifiers become out of date. To help reverse this trend, we converted BIND to a standard format, Proteomics Standard Initiative-Molecular Interaction 2.5, starting from the last curated data release (from 2005) available in a custom XML format and made the core components (interactions and complexes) plus additional valuable curated information available for download (http://download.baderlab.org/BINDTranslation/). Major work during the conversion process was required to update out of date molecule identifiers resulting in a more comprehensive conversion of BIND, by measures including number of species and interactor types covered, than what is currently accessible elsewhere. This work also highlights issues of data modeling, controlled vocabulary adoption and data cleaning that can serve as a general case study on the future compatibility of interaction databases. Database URL: http://download.baderlab.org/BINDTranslation/
Figures
Similar articles
-
Encompassing new use cases - level 3.0 of the HUPO-PSI format for molecular interactions.BMC Bioinformatics. 2018 Apr 11;19(1):134. doi: 10.1186/s12859-018-2118-1. BMC Bioinformatics. 2018. PMID: 29642841 Free PMC article.
-
Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions.BMC Biol. 2007 Oct 9;5:44. doi: 10.1186/1741-7007-5-44. BMC Biol. 2007. PMID: 17925023 Free PMC article.
-
IntAct: an open source molecular interaction database.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D452-5. doi: 10.1093/nar/gkh052. Nucleic Acids Res. 2004. PMID: 14681455 Free PMC article.
-
The work of the Human Proteome Organisation's Proteomics Standards Initiative (HUPO PSI).OMICS. 2006 Summer;10(2):145-51. doi: 10.1089/omi.2006.10.145. OMICS. 2006. PMID: 16901219 Review.
-
Proteomics data exchange and storage: the need for common standards and public repositories.Methods Mol Biol. 2013;1007:317-33. doi: 10.1007/978-1-62703-392-3_14. Methods Mol Biol. 2013. PMID: 23666733 Review.
Cited by
-
Identification of potential and novel target genes in pituitary prolactinoma by bioinformatics analysis.AIMS Neurosci. 2021 Feb 7;8(2):254-283. doi: 10.3934/Neuroscience.2021014. eCollection 2021. AIMS Neurosci. 2021. PMID: 33709028 Free PMC article.
-
Merging and scoring molecular interactions utilising existing community standards: tools, use-cases and a case study.Database (Oxford). 2015 Feb 4;2015:bau131. doi: 10.1093/database/bau131. Print 2015. Database (Oxford). 2015. PMID: 25652942 Free PMC article.
-
UniHI 7: an enhanced database for retrieval and interactive analysis of human molecular interaction networks.Nucleic Acids Res. 2014 Jan;42(Database issue):D408-14. doi: 10.1093/nar/gkt1100. Epub 2013 Nov 8. Nucleic Acids Res. 2014. PMID: 24214987 Free PMC article.
-
Duplicates, redundancies and inconsistencies in the primary nucleotide databases: a descriptive study.Database (Oxford). 2017 Jan 10;2017:baw163. doi: 10.1093/database/baw163. Print 2017. Database (Oxford). 2017. PMID: 28077566 Free PMC article.
-
Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain.Elife. 2016 Feb 19;5:e10421. doi: 10.7554/eLife.10421. Elife. 2016. PMID: 26894958 Free PMC article.
References
-
- Hogue CW. The other side of staying out of a BIND. Nat. Biotechnol. 2007;25:971. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

