Tunable signal processing in synthetic MAP kinase cascades
- PMID: 21215374
- PMCID: PMC3035479
- DOI: 10.1016/j.cell.2010.12.014
Tunable signal processing in synthetic MAP kinase cascades
Abstract
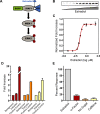
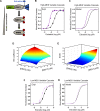
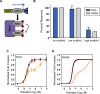
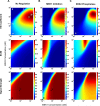
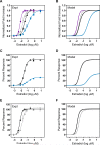
The flexibility of MAPK cascade responses enables regulation of a vast array of cell fate decisions, but elucidating the mechanisms underlying this plasticity is difficult in endogenous signaling networks. We constructed insulated mammalian MAPK cascades in yeast to explore how intrinsic and extrinsic perturbations affect the flexibility of these synthetic signaling modules. Contrary to biphasic dependence on scaffold concentration, we observe monotonic decreases in signal strength as scaffold concentration increases. We find that augmenting the concentration of sequential kinases can enhance ultrasensitivity and lower the activation threshold. Further, integrating negative regulation and concentration variation can decouple ultrasensitivity and threshold from the strength of the response. Computational analyses show that cascading can generate ultrasensitivity and that natural cascades with different kinase concentrations are innately biased toward their distinct activation profiles. This work demonstrates that tunable signal processing is inherent to minimal MAPK modules and elucidates principles for rational design of synthetic signaling systems.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Ultrasensitivity in signaling cascades revisited: Linking local and global ultrasensitivity estimations.PLoS One. 2017 Jun 29;12(6):e0180083. doi: 10.1371/journal.pone.0180083. eCollection 2017. PLoS One. 2017. PMID: 28662096 Free PMC article.
-
Simple synthetic protein scaffolds can create adjustable artificial MAPK circuits in yeast and mammalian cells.Sci Signal. 2015 Jun 30;8(383):ra66. doi: 10.1126/scisignal.aab3397. Sci Signal. 2015. PMID: 26126717
-
Ultrasensitivity part III: cascades, bistable switches, and oscillators.Trends Biochem Sci. 2014 Dec;39(12):612-8. doi: 10.1016/j.tibs.2014.10.002. Epub 2014 Nov 10. Trends Biochem Sci. 2014. PMID: 25456048 Free PMC article. Review.
-
Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades.Eur J Biochem. 2000 Mar;267(6):1583-8. doi: 10.1046/j.1432-1327.2000.01197.x. Eur J Biochem. 2000. PMID: 10712587
-
Scaffold proteins in mammalian MAP kinase cascades.J Biochem. 2004 Jun;135(6):657-61. doi: 10.1093/jb/mvh079. J Biochem. 2004. PMID: 15213240 Review.
Cited by
-
Ultrasensitive Negative Feedback Control: A Natural Approach for the Design of Synthetic Controllers.PLoS One. 2016 Aug 18;11(8):e0161605. doi: 10.1371/journal.pone.0161605. eCollection 2016. PLoS One. 2016. PMID: 27537373 Free PMC article.
-
Direct comparison of small RNA and transcription factor signaling.Nucleic Acids Res. 2012 Aug;40(15):7269-79. doi: 10.1093/nar/gks439. Epub 2012 May 22. Nucleic Acids Res. 2012. PMID: 22618873 Free PMC article.
-
Activation of extracellular signal-regulated kinase but not of p38 mitogen-activated protein kinase pathways in lymphocytes requires allosteric activation of SOS.Mol Cell Biol. 2013 Jun;33(12):2470-84. doi: 10.1128/MCB.01593-12. Epub 2013 Apr 15. Mol Cell Biol. 2013. PMID: 23589333 Free PMC article.
-
Spatial focalization of pheromone/MAPK signaling triggers commitment to cell-cell fusion.Genes Dev. 2016 Oct 1;30(19):2226-2239. doi: 10.1101/gad.286922.116. Genes Dev. 2016. PMID: 27798845 Free PMC article.
-
Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation.Sci Rep. 2014 Apr 15;4:4697. doi: 10.1038/srep04697. Sci Rep. 2014. PMID: 24732094 Free PMC article.
References
-
- Atienza J. Human ERK1 Induces Filamentous Growth and Cell Wall Remodeling Pathways in Saccharomyces cerevisiae. J Biol Chem. 2000;275:20638–20646. - PubMed
-
- Bagowski CP, Besser J, Frey CR, Ferrell JE., Jr. The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr Biol. 2003;13:315–320. - PubMed
-
- Bashor C, Helman N, Yan S, Lim W. Using Engineered Scaffold Interactions to Reshape MAP Kinase Pathway Signaling Dynamics. Science. 2008;319:1539–1543. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

