Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1
- PMID: 21203485
- PMCID: PMC3009601
- DOI: 10.1371/journal.ppat.1001236
Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1
Abstract
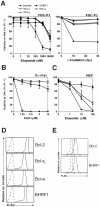
Epstein-Barr virus (EBV) is associated with human malignancies, especially those affecting the B cell compartment such as Burkitt lymphoma. The virally encoded homolog of the mammalian pro-survival protein Bcl-2, BHRF1 contributes to viral infectivity and lymphomagenesis. In addition to the pro-apoptotic BH3-only protein Bim, its key target in lymphoid cells, BHRF1 also binds a selective sub-set of pro-apoptotic proteins (Bid, Puma, Bak) expressed by host cells. A consequence of BHRF1 expression is marked resistance to a range of cytotoxic agents and in particular, we show that its expression renders a mouse model of Burkitt lymphoma untreatable. As current small organic antagonists of Bcl-2 do not target BHRF1, the structures of it in complex with Bim or Bak shown here will be useful to guide efforts to target BHRF1 in EBV-associated malignancies, which are usually associated with poor clinical outcomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
EBV BCL-2 homologue BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins.Cell Death Differ. 2020 May;27(5):1554-1568. doi: 10.1038/s41418-019-0435-1. Epub 2019 Oct 23. Cell Death Differ. 2020. PMID: 31645677 Free PMC article.
-
Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.Viruses. 2022 Oct 9;14(10):2222. doi: 10.3390/v14102222. Viruses. 2022. PMID: 36298777 Free PMC article.
-
The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim.Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5663-8. doi: 10.1073/pnas.0901036106. Epub 2009 Mar 17. Proc Natl Acad Sci U S A. 2009. PMID: 19293378 Free PMC article.
-
[Anti-apoptotic function of the Epstein-Barr virus LMP1 and BHRF1 proteins].Nihon Rinsho. 1996 Jul;54(7):1848-54. Nihon Rinsho. 1996. PMID: 8741677 Review. Japanese.
-
Burkitt lymphoma--a stalking horse for cancer research?Semin Cancer Biol. 2009 Dec;19(6):347-50. doi: 10.1016/j.semcancer.2009.07.001. Epub 2009 Jul 14. Semin Cancer Biol. 2009. PMID: 19607918 Review.
Cited by
-
Epstein-Barr virus: the mastermind of immune chaos.Front Immunol. 2024 Feb 7;15:1297994. doi: 10.3389/fimmu.2024.1297994. eCollection 2024. Front Immunol. 2024. PMID: 38384471 Free PMC article. Review.
-
Disruption of Bcl-2 and Bcl-xL by viral proteins as a possible cause of cancer.Infect Agent Cancer. 2014 Dec 23;9:44. doi: 10.1186/1750-9378-9-44. eCollection 2014. Infect Agent Cancer. 2014. PMID: 25699089 Free PMC article. Review.
-
Epstein-Barr virus evades CD4+ T cell responses in lytic cycle through BZLF1-mediated downregulation of CD74 and the cooperation of vBcl-2.PLoS Pathog. 2011 Dec;7(12):e1002455. doi: 10.1371/journal.ppat.1002455. Epub 2011 Dec 22. PLoS Pathog. 2011. PMID: 22216005 Free PMC article.
-
Essential role of hyperacetylated microtubules in innate immunity escape orchestrated by the EBV-encoded BHRF1 protein.PLoS Pathog. 2022 Mar 11;18(3):e1010371. doi: 10.1371/journal.ppat.1010371. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35275978 Free PMC article.
-
EBV BCL-2 homologue BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins.Cell Death Differ. 2020 May;27(5):1554-1568. doi: 10.1038/s41418-019-0435-1. Epub 2019 Oct 23. Cell Death Differ. 2020. PMID: 31645677 Free PMC article.
References
-
- Cuconati A, White E. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes and Development. 2002;16:2465–2478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

