Hexagonal assembly of a restricting TRIM5alpha protein
- PMID: 21187419
- PMCID: PMC3021009
- DOI: 10.1073/pnas.1013426108
Hexagonal assembly of a restricting TRIM5alpha protein
Abstract
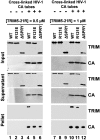
TRIM5α proteins are restriction factors that protect mammalian cells from retroviral infections by binding incoming viral capsids, accelerating their dissociation, and preventing reverse transcription of the viral genome. Individual TRIM5 isoforms can often protect cells against a broad range of retroviruses, as exemplified by rhesus monkey TRIM5α and its variant, TRIM5-21R, which recognize HIV-1 as well as several distantly related retroviruses. Although capsid recognition is not yet fully understood, previous work has shown that the C-terminal SPRY/B30.2 domain of dimeric TRIM5α binds directly to viral capsids, and that higher-order TRIM5α oligomerization appears to contribute to the efficiency of capsid recognition. Here, we report that recombinant TRIM5-21R spontaneously assembled into two-dimensional paracrystalline hexagonal lattices comprising open, six-sided rings. TRIM5-21R assembly did not require the C-terminal SPRY domain, but did require both protein dimerization and a B-box 2 residue (Arg121) previously implicated in TRIM5α restriction and higher-order assembly. Furthermore, TRIM5-21R assembly was promoted by binding to hexagonal arrays of the HIV-1 CA protein that mimic the surface of the viral capsid. We therefore propose that TRIM5α proteins have evolved to restrict a range of different retroviruses by assembling a deformable hexagonal scaffold that positions the capsid-binding domains to match the symmetry and spacing of the capsid surface lattice. Capsid recognition therefore involves a synergistic combination of direct binding interactions, avidity effects, templated assembly, and lattice complementarity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article.
-
Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication.J Virol. 2008 Dec;82(23):11682-94. doi: 10.1128/JVI.01562-08. Epub 2008 Sep 17. J Virol. 2008. PMID: 18799573 Free PMC article.
-
The Three-Fold Axis of the HIV-1 Capsid Lattice Is the Species-Specific Binding Interface for TRIM5α.J Virol. 2018 Feb 12;92(5):e01541-17. doi: 10.1128/JVI.01541-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237846 Free PMC article.
-
Recent insights into the mechanism and consequences of TRIM5α retroviral restriction.AIDS Res Hum Retroviruses. 2011 Mar;27(3):231-8. doi: 10.1089/AID.2010.0367. AIDS Res Hum Retroviruses. 2011. PMID: 21247355 Free PMC article. Review.
-
Anti-retroviral activity of TRIM5 alpha.Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review.
Cited by
-
A comparison of murine leukemia viruses that escape from human and rhesus macaque TRIM5αs.J Virol. 2013 Jun;87(11):6455-68. doi: 10.1128/JVI.03425-12. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536686 Free PMC article.
-
HIV suppression by host restriction factors and viral immune evasion.Curr Opin Struct Biol. 2015 Apr;31:106-14. doi: 10.1016/j.sbi.2015.04.004. Epub 2015 May 16. Curr Opin Struct Biol. 2015. PMID: 25939065 Free PMC article. Review.
-
Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers.Sci Rep. 2015 Jun 4;5:10819. doi: 10.1038/srep10819. Sci Rep. 2015. PMID: 26043233 Free PMC article.
-
More than one way to TRIM a capsid.Proc Natl Acad Sci U S A. 2012 Nov 27;109(48):19517-8. doi: 10.1073/pnas.1217596109. Epub 2012 Nov 13. Proc Natl Acad Sci U S A. 2012. PMID: 23150592 Free PMC article. No abstract available.
-
TRIM5α Restricts Flavivirus Replication by Targeting the Viral Protease for Proteasomal Degradation.Cell Rep. 2019 Jun 11;27(11):3269-3283.e6. doi: 10.1016/j.celrep.2019.05.040. Cell Rep. 2019. PMID: 31189110 Free PMC article.
References
-
- Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1G20RR031199/RR/NCRR NIH HHS/United States
- R37 AI045405-13/AI/NIAID NIH HHS/United States
- R01 GM066087-09/GM/NIGMS NIH HHS/United States
- R37 AI045405/AI/NIAID NIH HHS/United States
- P41 RR017573/RR/NCRR NIH HHS/United States
- R01-AI63987/AI/NIAID NIH HHS/United States
- 1S10RR025067/RR/NCRR NIH HHS/United States
- P50 GM082545/GM/NIGMS NIH HHS/United States
- R01 AI063987/AI/NIAID NIH HHS/United States
- P41 RR17573/RR/NCRR NIH HHS/United States
- P50-GM082545/GM/NIGMS NIH HHS/United States
- G20 RR031199/RR/NCRR NIH HHS/United States
- R01-GM066087/GM/NIGMS NIH HHS/United States
- R37 AI-45405-06/AI/NIAID NIH HHS/United States
- P50 GM082545-05/GM/NIGMS NIH HHS/United States
- S10 RR025067/RR/NCRR NIH HHS/United States
- R01 GM066087/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

