Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA
- PMID: 21185869
- PMCID: PMC3049850
- DOI: 10.1016/j.jviromet.2010.12.014
Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA
Abstract
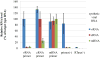
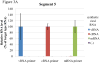
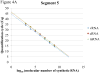
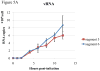
Real-time RT-PCR is used to quantify individual influenza viral RNAs. However, conventional real-time RT-PCR, using strand-specific primers, has been shown to produce not only the anticipated strand-specific products, but also substantial amounts of non-strand-specific products, indicating lack of specificity. Therefore, in this study, a novel strand-specific real-time RT-PCR method was established to quantify the three types of influenza viral RNA (vRNA, cRNA, and mRNA) separately. This method is based on reverse transcription using tagged primers to add a 'tag' sequence at the 5' end and the hot-start method. Real-time PCR using the 'tag' portion as the forward primer and a segment-specific reverse primer ensured the specificity for quantifying the three types of RNA. Using this method, specific target RNA was detected at 100-100,000-folds higher level than other types of RNA. This method was also used to evaluate the vRNA, cRNA, and mRNA levels of segments 5 and 6 in MDCK cells infected with influenza A virus at different time point post-infections. The cRNA level was 1/10 to 1/100 lower than that of the vRNA and mRNA. Moreover, different dynamics of vRNA, cRNA, and mRNA synthesis were observed; the copy number of the vRNA gradually increased throughout the infection, the cRNA increased and then plateaued, while the mRNA increased and then decreased. This novel method thus provides data critical for understanding the influenza virus life cycle, including transcription, replication, and genome incorporation into virions.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
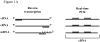
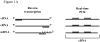




Similar articles
-
Development of strand-specific real-time RT-PCR for the analysis of SCRV transcription and replication dynamics.Microb Pathog. 2019 Apr;129:146-151. doi: 10.1016/j.micpath.2019.02.001. Epub 2019 Feb 4. Microb Pathog. 2019. PMID: 30731189
-
Development of strand-specific real-time RT-PCR to distinguish viral RNAs during Newcastle disease virus infection.ScientificWorldJournal. 2014;2014:934851. doi: 10.1155/2014/934851. Epub 2014 Oct 14. ScientificWorldJournal. 2014. PMID: 25379553 Free PMC article.
-
Transcription and replication of the influenza a virus genome.Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review.
-
Determination of the vRNA and cRNA promoter activity by M segment-specific non-coding nucleotides of influenza A virus.RNA Biol. 2021 May;18(5):785-795. doi: 10.1080/15476286.2020.1864182. Epub 2020 Dec 23. RNA Biol. 2021. PMID: 33317417 Free PMC article.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
Cited by
-
Eukaryotic Translation Elongation Factor 1 Delta Inhibits the Nuclear Import of the Nucleoprotein and PA-PB1 Heterodimer of Influenza A Virus.J Virol. 2020 Dec 22;95(2):e01391-20. doi: 10.1128/JVI.01391-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087462 Free PMC article.
-
ATG9A regulates the dissociation of recycling endosomes from microtubules to form liquid influenza A virus inclusions.PLoS Biol. 2023 Nov 20;21(11):e3002290. doi: 10.1371/journal.pbio.3002290. eCollection 2023 Nov. PLoS Biol. 2023. PMID: 37983294 Free PMC article.
-
Ebola virus-mediated T-lymphocyte depletion is the result of an abortive infection.PLoS Pathog. 2019 Oct 24;15(10):e1008068. doi: 10.1371/journal.ppat.1008068. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31648236 Free PMC article.
-
Avian Influenza A Virus Polymerase Recruits Cellular RNA Helicase eIF4A3 to Promote Viral mRNA Splicing and Spliced mRNA Nuclear Export.Front Microbiol. 2019 Jul 16;10:1625. doi: 10.3389/fmicb.2019.01625. eCollection 2019. Front Microbiol. 2019. PMID: 31379779 Free PMC article.
-
Hemagglutinin Stability Regulates H1N1 Influenza Virus Replication and Pathogenicity in Mice by Modulating Type I Interferon Responses in Dendritic Cells.J Virol. 2020 Jan 17;94(3):e01423-19. doi: 10.1128/JVI.01423-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694942 Free PMC article.
References
-
- Craggs J, Ball J, Thomson B, Irving W, Grabowska A. Development of a strand-specific RT-PCR based assay to detect the replicative form of hepatitis C virus RNA. J Virol Methods. 2001;94:111–120. - PubMed
-
- Hatada E, Hasegawa M, Mukaigawa J, Shimizu K, Fukuda R. Control of influenza virus gene expression: quantitative analysis of each viral RNA species in infected cells. J Biochem. 1989;105:537–546. - PubMed
-
- Karlas A, Machuy N, Shin Y, Pleissner K, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie L, Hess S, Mäurer A, Müller E, Wolff T, Rudel T, Meyer T. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

