Dopamine D2 and D4 receptor heteromerization and its allosteric receptor-receptor interactions
- PMID: 21184734
- PMCID: PMC9479170
- DOI: 10.1016/j.bbrc.2010.12.083
Dopamine D2 and D4 receptor heteromerization and its allosteric receptor-receptor interactions
Abstract
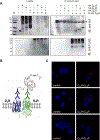
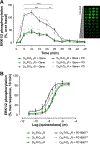
Dopamine D(2) and D(4) receptors partially codistribute in the dorsal striatum and appear to play a fundamental role in complex behaviors and motor function. The discovery of D(2)R-D(4.)(x)R (D(4.2)R, D(4.4)R or D(4.7)R) heteromers has been made in cellular models using co-immunoprecipitation, in situ Proximity Ligation Assays and BRET(1) techniques with the D(2)R and D(4.7)R receptors being the least effective in forming heteromers. Allosteric receptor-receptor interactions in D(2)R-D(4.2)R and D(2)R-D(4.4) R heteromers were observed using the MAPK assays indicating the existence of an enhancing allosteric receptor-receptor interaction in the corresponding heteromers between the two orthosteric binding sites. The bioinformatic predictions suggest the existence of a basic set of common triplets (ALQ and LRA) in the two participating receptors that may contribute to the receptor-receptor interaction interfaces.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Dopamine D2 receptor signaling dynamics of dopamine D2-neurotensin 1 receptor heteromers.Biochem Biophys Res Commun. 2013 May 24;435(1):140-6. doi: 10.1016/j.bbrc.2013.04.058. Epub 2013 Apr 25. Biochem Biophys Res Commun. 2013. PMID: 23624386
-
Dopamine D2 and angiotensin II type 1 receptors form functional heteromers in rat striatum.Biochem Pharmacol. 2015 Jul 15;96(2):131-42. doi: 10.1016/j.bcp.2015.05.006. Epub 2015 May 16. Biochem Pharmacol. 2015. PMID: 25986885
-
Characterization of the A2AR-D2R interface: focus on the role of the C-terminal tail and the transmembrane helices.Biochem Biophys Res Commun. 2010 Nov 26;402(4):801-7. doi: 10.1016/j.bbrc.2010.10.122. Epub 2010 Oct 30. Biochem Biophys Res Commun. 2010. PMID: 21040702
-
Moonlighting characteristics of G protein-coupled receptors: focus on receptor heteromers and relevance for neurodegeneration.IUBMB Life. 2011 Jul;63(7):463-72. doi: 10.1002/iub.473. IUBMB Life. 2011. PMID: 21698749 Review.
-
Molecular integration via allosteric interactions in receptor heteromers. A working hypothesis.Curr Opin Pharmacol. 2010 Feb;10(1):14-22. doi: 10.1016/j.coph.2009.10.010. Epub 2009 Nov 26. Curr Opin Pharmacol. 2010. PMID: 19942481 Review.
Cited by
-
On the existence and function of galanin receptor heteromers in the central nervous system.Front Endocrinol (Lausanne). 2012 Oct 26;3:127. doi: 10.3389/fendo.2012.00127. eCollection 2012. Front Endocrinol (Lausanne). 2012. PMID: 23112793 Free PMC article.
-
Heterodimerization of Mu Opioid Receptor Protomer with Dopamine D2 Receptor Modulates Agonist-Induced Internalization of Mu Opioid Receptor.Biomolecules. 2019 Aug 14;9(8):368. doi: 10.3390/biom9080368. Biomolecules. 2019. PMID: 31416253 Free PMC article.
-
Post-ER Stress Biogenesis of Golgi Is Governed by Giantin.Cells. 2019 Dec 13;8(12):1631. doi: 10.3390/cells8121631. Cells. 2019. PMID: 31847122 Free PMC article.
-
Disruption of A2AR-D2R Heteroreceptor Complexes After A2AR Transmembrane 5 Peptide Administration Enhances Cocaine Self-Administration in Rats.Mol Neurobiol. 2018 Aug;55(8):7038-7048. doi: 10.1007/s12035-018-0887-1. Epub 2018 Jan 30. Mol Neurobiol. 2018. PMID: 29383683 Free PMC article.
-
Approaching "phantom heritability" in psychiatry by hypothesis-driven gene-gene interactions.Front Hum Neurosci. 2013 May 16;7:210. doi: 10.3389/fnhum.2013.00210. eCollection 2013. Front Hum Neurosci. 2013. PMID: 23720624 Free PMC article. No abstract available.
References
-
- Scarselli M, Novi F, Schallmach E, Lin R, Baragli A, Colzi A, Griffon N, Corsini GU, Sokoloff P, Levenson R, Vogel Z, Maggio R, D2/D3 dopamine receptor heterodimers exhibit unique functional properties, J. Biol. Chem 276 (2001) 30308–30314. - PubMed
-
- Rashid AJ, O’Dowd BF, Verma V, George SR, Neuronal Gq/11-coupled dopamine receptors: an uncharted role for dopamine, Trends Pharmacol. Sci 28 (2007) 551–555. - PubMed
-
- Rubinstein M, Phillips TJ, Bunzow JR, Falzone TL, Dziewczapolski G, Zhang G, Fang Y, Larson JL, McDougall JA, Chester JA, Saez C, Pugsley TA, Gershanik O, Low MJ, Grandy DK, Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine, Cell 90 (1997) 991–1001. - PubMed
-
- Fuxe K, Ferre S, Canals M, Torvinen M, Terasmaa A, Marcellino D, Goldberg SR, Staines W, Jacobsen KX, Lluis C, Woods AS, Agnati LF, Franco R, Adenosine A2A and dopamine D2 heteromeric receptor complexes and their function, J. Mol. Neurosci 26 (2005) 209–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

