Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate
- PMID: 21183466
- PMCID: PMC3082910
- DOI: 10.1093/nar/gkq1241
Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate
Abstract
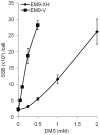
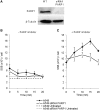
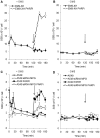
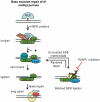
Base excision repair (BER) represents the most important repair pathway of endogenous DNA lesions. Initially, a base damage is recognized, excised and a DNA single-strand break (SSB) intermediate forms. The SSB is then ligated, a process that employs proteins also involved in SSB repair, e.g. XRCC1, Ligase III and possibly PARP1. Here, we confirm the role of XRCC1 and PARP in direct SSB repair. Interestingly, we uncover a synthetic lethality between XRCC1 deficiency and PARP inhibition. We also treated cells with alkylating agent dimethyl sulfate (DMS) and monitored the SSB intermediates formed during BER. DMS-induced SSBs were quickly repaired in wild-type cells; while a rapid accumulation of SSBs was observed in cells where post-incision repair was blocked by a PARP inhibitor or by XRCC1 deficiency (EM9 cells). Interestingly, DMS-induced SSBs did not accumulate in PARP1 siRNA depleted cells, demonstrating that PARP1 is not required for efficient completion of BER. Based on these results we suggest no immediate role for PARP1 in BER, but that PARP inhibitors trap PARP on the SSB intermediate formed during BER. Unexpectedly, addition of PARP inhibitor 2 h after DMS treatment still increased SSB levels indicating ongoing repair even at this late time point.
Figures



Similar articles
-
XRCC1 counteracts poly(ADP ribose)polymerase (PARP) poisons, olaparib and talazoparib, and a clinical alkylating agent, temozolomide, by promoting the removal of trapped PARP1 from broken DNA.Genes Cells. 2022 May;27(5):331-344. doi: 10.1111/gtc.12929. Epub 2022 Mar 1. Genes Cells. 2022. PMID: 35194903 Free PMC article.
-
XRCC1 prevents toxic PARP1 trapping during DNA base excision repair.Mol Cell. 2021 Jul 15;81(14):3018-3030.e5. doi: 10.1016/j.molcel.2021.05.009. Epub 2021 Jun 7. Mol Cell. 2021. PMID: 34102106 Free PMC article.
-
CK2 phosphorylation of XRCC1 facilitates dissociation from DNA and single-strand break formation during base excision repair.DNA Repair (Amst). 2011 Sep 5;10(9):961-9. doi: 10.1016/j.dnarep.2011.07.004. Epub 2011 Aug 15. DNA Repair (Amst). 2011. PMID: 21840775
-
XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks.Cell Res. 2008 Jan;18(1):48-63. doi: 10.1038/cr.2008.7. Cell Res. 2008. PMID: 18166976 Free PMC article. Review.
-
Completion of base excision repair by mammalian DNA ligases.Prog Nucleic Acid Res Mol Biol. 2001;68:151-64. doi: 10.1016/s0079-6603(01)68097-8. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554294 Review.
Cited by
-
NEO212, temozolomide conjugated to NEO100, exerts superior therapeutic activity over temozolomide in preclinical chemoradiation models of glioblastoma.Neurooncol Adv. 2024 Jun 11;6(1):vdae095. doi: 10.1093/noajnl/vdae095. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 39022643 Free PMC article.
-
Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1.PLoS Genet. 2013 Apr;9(4):e1003413. doi: 10.1371/journal.pgen.1003413. Epub 2013 Apr 4. PLoS Genet. 2013. PMID: 23593019 Free PMC article.
-
Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions.Nucleic Acids Res. 2015 Apr 30;43(8):4028-38. doi: 10.1093/nar/gkv250. Epub 2015 Mar 26. Nucleic Acids Res. 2015. PMID: 25813046 Free PMC article.
-
The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy.Cells. 2022 Nov 27;11(23):3798. doi: 10.3390/cells11233798. Cells. 2022. PMID: 36497058 Free PMC article. Review.
-
TDP1 deficiency sensitizes human cells to base damage via distinct topoisomerase I and PARP mechanisms with potential applications for cancer therapy.Nucleic Acids Res. 2014 Mar;42(5):3089-103. doi: 10.1093/nar/gkt1260. Epub 2013 Dec 13. Nucleic Acids Res. 2014. PMID: 24335147 Free PMC article.
References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. - PubMed
-
- Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

