Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins
- PMID: 21182249
- PMCID: PMC3117330
- DOI: 10.1021/ja109581t
Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins
Abstract
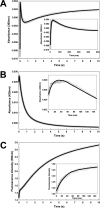
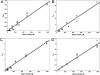
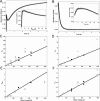
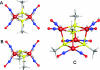
The reactivity of protein bound iron-sulfur clusters with nitric oxide (NO) is well documented, but little is known about the actual mechanism of cluster nitrosylation. Here, we report studies of members of the Wbl family of [4Fe-4S] containing proteins, which play key roles in regulating developmental processes in actinomycetes, including Streptomyces and Mycobacteria, and have been shown to be NO responsive. Streptomyces coelicolor WhiD and Mycobacterium tuberculosis WhiB1 react extremely rapidly with NO in a multiphasic reaction involving, remarkably, 8 NO molecules per [4Fe-4S] cluster. The reaction is 10(4)-fold faster than that observed with O(2) and is by far the most rapid iron-sulfur cluster nitrosylation reaction reported to date. An overall stoichiometry of [Fe(4)S(4)(Cys)(4)](2-) + 8NO → 2[Fe(I)(2)(NO)(4)(Cys)(2)](0) + S(2-) + 3S(0) has been established by determination of the sulfur products and their oxidation states. Kinetic analysis leads to a four-step mechanism that accounts for the observed NO dependence. DFT calculations suggest the possibility that the nitrosylation product is a novel cluster [Fe(I)(4)(NO)(8)(Cys)(4)](0) derived by dimerization of a pair of Roussin's red ester (RRE) complexes.
Figures


Similar articles
-
Mass Spectrometric Identification of [4Fe-4S](NO)x Intermediates of Nitric Oxide Sensing by Regulatory Iron-Sulfur Cluster Proteins.Chemistry. 2019 Mar 7;25(14):3675-3684. doi: 10.1002/chem.201806113. Epub 2019 Feb 7. Chemistry. 2019. PMID: 30600851
-
Mechanism of [4Fe-4S](Cys)4 cluster nitrosylation is conserved among NO-responsive regulators.J Biol Chem. 2013 Apr 19;288(16):11492-502. doi: 10.1074/jbc.M112.439901. Epub 2013 Mar 7. J Biol Chem. 2013. PMID: 23471974 Free PMC article.
-
Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide.Acc Chem Res. 2014 Oct 21;47(10):3196-205. doi: 10.1021/ar5002507. Epub 2014 Sep 29. Acc Chem Res. 2014. PMID: 25262769 Review.
-
Interaction of the Streptomyces Wbl protein WhiD with the principal sigma factor σHrdB depends on the WhiD [4Fe-4S] cluster.J Biol Chem. 2020 Jul 10;295(28):9752-9765. doi: 10.1074/jbc.RA120.012708. Epub 2020 Apr 16. J Biol Chem. 2020. PMID: 32303639 Free PMC article.
-
WhiB-like proteins: Diversity of structure, function and mechanism.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119787. doi: 10.1016/j.bbamcr.2024.119787. Epub 2024 Jun 13. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38879133 Review.
Cited by
-
Sensing mechanisms of iron-sulfur cluster regulatory proteins elucidated using native mass spectrometry.Dalton Trans. 2021 Jun 21;50(23):7887-7897. doi: 10.1039/d1dt00993a. Epub 2021 May 25. Dalton Trans. 2021. PMID: 34037038 Free PMC article.
-
Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2.Tuberculosis (Edinb). 2012 Jul;92(4):328-32. doi: 10.1016/j.tube.2012.03.001. Epub 2012 Mar 29. Tuberculosis (Edinb). 2012. PMID: 22464736 Free PMC article.
-
Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae.mBio. 2013 Sep 24;4(5):e00684-13. doi: 10.1128/mBio.00684-13. mBio. 2013. PMID: 24065632 Free PMC article.
-
Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria.Nucleic Acids Res. 2015 Jun 23;43(11):5377-93. doi: 10.1093/nar/gkv420. Epub 2015 May 4. Nucleic Acids Res. 2015. PMID: 25940627 Free PMC article.
-
Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence.Antioxid Redox Signal. 2012 Apr 1;16(7):687-97. doi: 10.1089/ars.2011.4341. Antioxid Redox Signal. 2012. PMID: 22010944 Free PMC article. Review.
References
-
- Lancaster J. R. Jr. Nitric Oxide 1997, 1, 18–30. - PubMed
-
- Cooper C. E. Biochim. Biophys. Acta 1999, 1411, 290–309. - PubMed
-
- Bruckdorfer R. Mol. Aspects Med. 2005, 26, 3–31. - PubMed
-
- Watmough N. J.; Butland G.; Cheesman M. R.; Moir J. W.; Richardson D. J.; Spiro S. Biochim. Biophys. Acta 1999, 1411, 456–474. - PubMed
-
- Poole R. K. Biochem. Soc. Trans. 2005, 33, 176–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 078731/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- BB/D00926X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U117585867/MRC_/Medical Research Council/United Kingdom
- BB/G019347/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000015/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

