Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription
- PMID: 21160280
- PMCID: PMC3073334
- DOI: 10.4161/rna.7.6.14115
Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription
Abstract
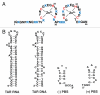
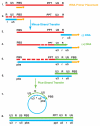
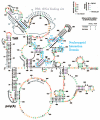
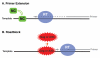
The HIV-1 nucleocapsid protein (NC) is a nucleic acid chaperone, which remodels nucleic acid structures so that the most thermodynamically stable conformations are formed. This activity is essential for virus replication and has a critical role in mediating highly specific and efficient reverse transcription. NC's function in this process depends upon three properties: (1) ability to aggregate nucleic acids; (2) moderate duplex destabilization activity; and (3) rapid on-off binding kinetics. Here, we present a detailed molecular analysis of the individual events that occur during viral DNA synthesis and show how NC's properties are important for almost every step in the pathway. Finally, we also review biological aspects of reverse transcription during infection and the interplay between NC, reverse transcriptase, and human APOBEC3G, an HIV-1 restriction factor that inhibits reverse transcription and virus replication in the absence of the HIV-1 Vif protein.
Figures
Similar articles
-
Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein's nucleic acid chaperone activity and retroviral replication.Virus Res. 2013 Feb;171(2):263-77. doi: 10.1016/j.virusres.2012.07.008. Epub 2012 Jul 16. Virus Res. 2013. PMID: 22814429 Free PMC article.
-
Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G.Nucleic Acids Res. 2007;35(21):7096-108. doi: 10.1093/nar/gkm750. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17942420 Free PMC article.
-
Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function.J Mol Biol. 2006 Nov 10;363(5):867-77. doi: 10.1016/j.jmb.2006.08.070. Epub 2006 Aug 30. J Mol Biol. 2006. PMID: 16997322
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Nucleocapsid protein function in early infection processes.Virus Res. 2008 Jun;134(1-2):39-63. doi: 10.1016/j.virusres.2007.12.006. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279991 Free PMC article. Review.
Cited by
-
Moloney murine leukemia virus genomic RNA packaged in the absence of a full complement of wild type nucleocapsid protein.Virology. 2012 Sep 1;430(2):100-9. doi: 10.1016/j.virol.2012.05.003. Epub 2012 May 25. Virology. 2012. PMID: 22633243 Free PMC article.
-
Nucleocapsid Protein Precursors NCp9 and NCp15 Suppress ATP-Mediated Rescue of AZT-Terminated Primers by HIV-1 Reverse Transcriptase.Antimicrob Agents Chemother. 2020 Sep 21;64(10):e00958-20. doi: 10.1128/AAC.00958-20. Print 2020 Sep 21. Antimicrob Agents Chemother. 2020. PMID: 32747359 Free PMC article.
-
Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities.Retrovirology. 2015 Jan 22;12:3. doi: 10.1186/s12977-014-0130-8. Retrovirology. 2015. PMID: 25614027 Free PMC article.
-
Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties.Nucleic Acids Res. 2014 Jan;42(2):1095-110. doi: 10.1093/nar/gkt945. Epub 2013 Oct 24. Nucleic Acids Res. 2014. PMID: 24163103 Free PMC article.
-
HIV-1 reverse transcriptase dissociates during strand transfer.J Mol Biol. 2011 Sep 23;412(3):354-64. doi: 10.1016/j.jmb.2011.07.055. Epub 2011 Jul 29. J Mol Biol. 2011. PMID: 21821047 Free PMC article.
References
-
- Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. - PubMed
-
- Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. - PubMed
-
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. - PubMed
-
- Henderson LE, Bowers MA, Sowder RC, II, Serabyn SA, Johnson DG, Bess JW, Jr, et al. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings and complete amino acid sequences. J Virol. 1992;66:1856–1865. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
