Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine
- PMID: 21151123
- PMCID: PMC3107705
- DOI: 10.1038/nbt.1732
Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine
Abstract
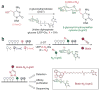
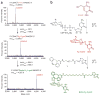
In contrast to 5-methylcytosine (5-mC), which has been studied extensively, little is known about 5-hydroxymethylcytosine (5-hmC), a recently identified epigenetic modification present in substantial amounts in certain mammalian cell types. Here we present a method for determining the genome-wide distribution of 5-hmC. We use the T4 bacteriophage β-glucosyltransferase to transfer an engineered glucose moiety containing an azide group onto the hydroxyl group of 5-hmC. The azide group can be chemically modified with biotin for detection, affinity enrichment and sequencing of 5-hmC-containing DNA fragments in mammalian genomes. Using this method, we demonstrate that 5-hmC is present in human cell lines beyond those previously recognized. We also find a gene expression level-dependent enrichment of intragenic 5-hmC in mouse cerebellum and an age-dependent acquisition of this modification in specific gene bodies linked to neurodegenerative disorders.
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at
Figures
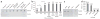
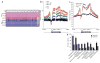
Comment in
-
Profiling the sixth base.Nat Methods. 2011 Jul;8(7):532-3. doi: 10.1038/nmeth0711-532b. Nat Methods. 2011. PMID: 21850733 No abstract available.
Similar articles
-
Bioorthogonal labeling of 5-hydroxymethylcytosine in genomic DNA and diazirine-based DNA photo-cross-linking probes.Acc Chem Res. 2011 Sep 20;44(9):709-17. doi: 10.1021/ar2000502. Epub 2011 May 3. Acc Chem Res. 2011. PMID: 21539303 Free PMC article.
-
Selective capture of 5-hydroxymethylcytosine from genomic DNA.J Vis Exp. 2012 Oct 5;(68):4441. doi: 10.3791/4441. J Vis Exp. 2012. PMID: 23070273 Free PMC article.
-
Biochemical characterization of recombinant β-glucosyltransferase and analysis of global 5-hydroxymethylcytosine in unique genomes.Biochemistry. 2012 Feb 7;51(5):1009-19. doi: 10.1021/bi2014739. Epub 2012 Jan 27. Biochemistry. 2012. PMID: 22229759 Free PMC article.
-
5-Hydroxymethylcytosine, the sixth base of the genome.Angew Chem Int Ed Engl. 2011 Jul 11;50(29):6460-8. doi: 10.1002/anie.201101547. Epub 2011 Jun 17. Angew Chem Int Ed Engl. 2011. PMID: 21688365 Review.
-
High-throughput sequencing offers new insights into 5-hydroxymethylcytosine.Biomol Concepts. 2016 Jun 1;7(3):169-78. doi: 10.1515/bmc-2016-0011. Biomol Concepts. 2016. PMID: 27356236 Free PMC article. Review.
Cited by
-
5-Hydroxymethylation alterations in cell-free DNA reflect molecular distinctions of diffuse large B cell lymphoma at different primary sites.Clin Epigenetics. 2022 Oct 11;14(1):126. doi: 10.1186/s13148-022-01344-1. Clin Epigenetics. 2022. PMID: 36221115 Free PMC article.
-
Distinctive Patterns of 5-Methylcytosine and 5-Hydroxymethylcytosine in Schizophrenia.Int J Mol Sci. 2024 Jan 4;25(1):636. doi: 10.3390/ijms25010636. Int J Mol Sci. 2024. PMID: 38203806 Free PMC article. Review.
-
Chemical reporters for biological discovery.Nat Chem Biol. 2013 Aug;9(8):475-84. doi: 10.1038/nchembio.1296. Nat Chem Biol. 2013. PMID: 23868317 Free PMC article. Review.
-
Genome-wide antagonism between 5-hydroxymethylcytosine and DNA methylation in the adult mouse brain.Front Biol (Beijing). 2014 Feb;9(1):66-74. doi: 10.1007/s11515-014-1295-1. Front Biol (Beijing). 2014. PMID: 25568643 Free PMC article.
-
Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction.Curr Alzheimer Res. 2012 Jun;9(5):536-44. doi: 10.2174/156720512800618035. Curr Alzheimer Res. 2012. PMID: 22272625 Free PMC article.
References
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. - PubMed
-
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
-
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R56 MH076090/MH/NIMH NIH HHS/United States
- R01 GM071440-05S1/GM/NIGMS NIH HHS/United States
- MH076090/MH/NIMH NIH HHS/United States
- R01 GM071440/GM/NIGMS NIH HHS/United States
- NS051630/NS/NINDS NIH HHS/United States
- R01 NS051630-05/NS/NINDS NIH HHS/United States
- MH078972/MH/NIMH NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R21 NS067461-01A1/NS/NINDS NIH HHS/United States
- R01 MH076090-05/MH/NIMH NIH HHS/United States
- R01 NS051630/NS/NINDS NIH HHS/United States
- R01 MH078972/MH/NIMH NIH HHS/United States
- R01 MH076090/MH/NIMH NIH HHS/United States
- R21 NS067461/NS/NINDS NIH HHS/United States
- GM071440/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

