Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes
- PMID: 21149566
- PMCID: PMC3002026
- DOI: 10.1083/jcb.201005046
Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes
Abstract
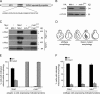
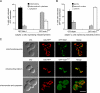
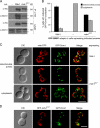
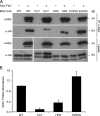
Recruitment and assembly of some dynamin-related guanosine triphosphatases depends on adaptor proteins restricted to distinct cellular membranes. The yeast Mdv1 adaptor localizes to mitochondria by binding to the membrane protein Fis1. Subsequent Mdv1 binding to the mitochondrial dynamin Dnm1 stimulates Dnm1 assembly into spirals, which encircle and divide the mitochondrial compartment. In this study, we report that dimeric Mdv1 is joined at its center by a 92-Å antiparallel coiled coil (CC). Modeling of the Fis1-Mdv1 complex using available crystal structures suggests that the Mdv1 CC lies parallel to the bilayer with N termini at opposite ends bound to Fis1 and C-terminal β-propeller domains (Dnm1-binding sites) extending into the cytoplasm. A CC length of appropriate length and sequence is necessary for optimal Mdv1 interaction with Fis1 and Dnm1 and is important for proper Dnm1 assembly before membrane scission. Our results provide a framework for understanding how adaptors act as scaffolds to orient and stabilize the assembly of dynamins on membranes.
Figures


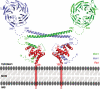
Similar articles
-
Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (Mdv1) coiled coil.J Biol Chem. 2012 Mar 23;287(13):9855-9861. doi: 10.1074/jbc.M111.329359. Epub 2012 Feb 1. J Biol Chem. 2012. PMID: 22303011 Free PMC article.
-
Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm.J Biol Chem. 2007 Nov 16;282(46):33769-33775. doi: 10.1074/jbc.M700807200. Epub 2007 Sep 20. J Biol Chem. 2007. PMID: 17884824 Free PMC article.
-
A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment.J Cell Biol. 2012 Nov 12;199(4):613-22. doi: 10.1083/jcb.201207079. J Cell Biol. 2012. PMID: 23148233 Free PMC article.
-
The machines that divide and fuse mitochondria.Annu Rev Biochem. 2007;76:751-80. doi: 10.1146/annurev.biochem.76.071905.090048. Annu Rev Biochem. 2007. PMID: 17362197 Review.
-
Dynamin assembly strategies and adaptor proteins in mitochondrial fission.Curr Biol. 2013 Oct 7;23(19):R891-9. doi: 10.1016/j.cub.2013.08.040. Curr Biol. 2013. PMID: 24112988 Free PMC article. Review.
Cited by
-
Biased placement of Mitochondria fission facilitates asymmetric inheritance of protein aggregates during yeast cell division.PLoS Comput Biol. 2023 Nov 27;19(11):e1011588. doi: 10.1371/journal.pcbi.1011588. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 38011208 Free PMC article.
-
Mff oligomerization is required for Drp1 activation and synergy with actin filaments during mitochondrial division.Mol Biol Cell. 2021 Oct 1;32(20):ar5. doi: 10.1091/mbc.E21-04-0224. Epub 2021 Aug 4. Mol Biol Cell. 2021. PMID: 34347505 Free PMC article.
-
Mitochondrial Inheritance in Phytopathogenic Fungi-Everything Is Known, or Is It?Int J Mol Sci. 2020 May 29;21(11):3883. doi: 10.3390/ijms21113883. Int J Mol Sci. 2020. PMID: 32485941 Free PMC article. Review.
-
Fis1 deficiencies differentially affect mitochondrial quality in skeletal muscle.Mitochondrion. 2019 Nov;49:217-226. doi: 10.1016/j.mito.2019.09.005. Epub 2019 Sep 14. Mitochondrion. 2019. PMID: 31526891 Free PMC article.
-
Structural Basis of Mitochondrial Scaffolds by Prohibitin Complexes: Insight into a Role of the Coiled-Coil Region.iScience. 2019 Sep 27;19:1065-1078. doi: 10.1016/j.isci.2019.08.056. Epub 2019 Sep 3. iScience. 2019. PMID: 31522117 Free PMC article.
References
-
- Collaborative Computational Project 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. D50:760–763 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

