Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes
- PMID: 21147927
- PMCID: PMC3028914
- DOI: 10.1128/JVI.02268-10
Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes
Abstract
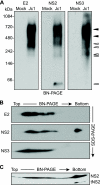
The hepatitis C virus (HCV) NS2 protein is essential for particle assembly, but its function in this process is unknown. We previously identified critical genetic interactions between NS2 and the viral E1-E2 glycoprotein and NS3-NS4A enzyme complexes. Based on these data, we hypothesized that interactions between these viral proteins are essential for HCV particle assembly. To identify interaction partners of NS2, we developed methods to site-specifically biotinylate NS2 in vivo and affinity capture NS2-containing protein complexes from virus-producing cells with streptavidin magnetic beads. By using these methods, we confirmed that NS2 physically interacts with E1, E2, and NS3 but did not stably interact with viral core or NS5A proteins. We further characterized these protein complexes by blue native polyacrylamide gel electrophoresis and identified ≈ 520-kDa and ≈ 680-kDa complexes containing E2, NS2, and NS3. The formation of NS2 protein complexes was dependent on coexpression of the viral p7 protein and enhanced by cotranslation of viral proteins as a polyprotein. Further characterization indicated that the glycoprotein complex interacts with NS2 via E2, and the pattern of N-linked glycosylation on E1 and E2 suggested that these interactions occur in the early secretory pathway. Importantly, several mutations that inhibited virus assembly were shown to inhibit NS2 protein complex formation, and NS2 was essential for mediating the interaction between E2 and NS3. These studies demonstrate that NS2 plays a central organizing role in HCV particle assembly by bringing together viral structural and nonstructural proteins.
Figures







Similar articles
-
Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes.J Virol. 2009 Sep;83(17):8379-95. doi: 10.1128/JVI.00891-09. Epub 2009 Jun 10. J Virol. 2009. PMID: 19515772 Free PMC article.
-
Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins.J Virol. 2011 Jan;85(1):86-97. doi: 10.1128/JVI.01070-10. Epub 2010 Oct 20. J Virol. 2011. PMID: 20962101 Free PMC article.
-
Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus.J Virol. 2007 Jan;81(2):629-38. doi: 10.1128/JVI.01890-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079282 Free PMC article.
-
Molecular virology of the hepatitis C virus.J Hepatol. 1999;31 Suppl 1:47-53. doi: 10.1016/s0168-8278(99)80374-2. J Hepatol. 1999. PMID: 10622560 Review.
-
Processing pathways of the hepatitis C virus proteins.J Hepatol. 1996;24(2 Suppl):11-9. J Hepatol. 1996. PMID: 8836884 Review.
Cited by
-
Signal peptidase complex subunit 1 participates in the assembly of hepatitis C virus through an interaction with E2 and NS2.PLoS Pathog. 2013;9(8):e1003589. doi: 10.1371/journal.ppat.1003589. Epub 2013 Aug 29. PLoS Pathog. 2013. PMID: 24009510 Free PMC article.
-
A Point Mutation in the N-Terminal Amphipathic Helix α0 in NS3 Promotes Hepatitis C Virus Assembly by Altering Core Localization to the Endoplasmic Reticulum and Facilitating Virus Budding.J Virol. 2017 Feb 28;91(6):e02399-16. doi: 10.1128/JVI.02399-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053108 Free PMC article.
-
Classical swine fever virus nonstructural protein p7 modulates infectious virus production.Sci Rep. 2017 Oct 11;7(1):12995. doi: 10.1038/s41598-017-13352-w. Sci Rep. 2017. PMID: 29021567 Free PMC article.
-
The intraviral protein interaction network of hepatitis C virus.Mol Cell Proteomics. 2014 Jul;13(7):1676-89. doi: 10.1074/mcp.M113.036301. Epub 2014 May 5. Mol Cell Proteomics. 2014. PMID: 24797426 Free PMC article.
-
Ultrastructural and biochemical basis for hepatitis C virus morphogenesis.Virus Genes. 2017 Apr;53(2):151-164. doi: 10.1007/s11262-017-1426-2. Epub 2017 Feb 23. Virus Genes. 2017. PMID: 28233195 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

