Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody
- PMID: 21134642
- PMCID: PMC3005625
- DOI: 10.1016/j.str.2010.09.017
Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody
Abstract
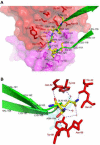
Ibalizumab is a humanized, anti-CD4 monoclonal antibody. It potently blocks HIV-1 infection and targets an epitope in the second domain of CD4 without interfering with immune functions mediated by interaction of CD4 with major histocompatibility complex (MHC) class II molecules. We report here the crystal structure of ibalizumab Fab fragment in complex with the first two domains (D1-D2) of CD4 at 2.2 Å resolution. Ibalizumab grips CD4 primarily by the BC-loop (residues 121-125) of D2, sitting on the opposite side of gp120 and MHC-II binding sites. No major conformational change in CD4 accompanies binding to ibalizumab. Both monovalent and bivalent forms of ibalizumab effectively block viral infection, suggesting that it does not need to crosslink CD4 to exert antiviral activity. While gp120-induced structural rearrangements in CD4 are probably minimal, CD4 structural rigidity is dispensable for ibalizumab inhibition. These results could guide CD4-based immunogen design and lead to a better understanding of HIV-1 entry.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Epitope mapping of ibalizumab, a humanized anti-CD4 monoclonal antibody with anti-HIV-1 activity in infected patients.J Virol. 2010 Jul;84(14):6935-42. doi: 10.1128/JVI.00453-10. Epub 2010 May 12. J Virol. 2010. PMID: 20463063 Free PMC article.
-
A non-canonical binding interface in the crystal structure of HIV-1 gp120 core in complex with CD4.Sci Rep. 2017 Apr 21;7:46733. doi: 10.1038/srep46733. Sci Rep. 2017. PMID: 28429756 Free PMC article.
-
Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker.J Virol. 2006 Jul;80(14):6725-37. doi: 10.1128/JVI.00118-06. J Virol. 2006. PMID: 16809278 Free PMC article.
-
Antiviral effects of CD4 derivatives.Curr Opin Immunol. 1989-1990 Feb;2(3):433-8. doi: 10.1016/0952-7915(89)90156-8. Curr Opin Immunol. 1989. PMID: 2576772 Review. No abstract available.
-
HIV gp120: double lock strategy foils host defences.Structure. 1998 Aug 15;6(8):945-9. doi: 10.1016/s0969-2126(98)00096-3. Structure. 1998. PMID: 9739096 Review.
Cited by
-
Distinct HIV-1 Neutralization Potency Profiles of Ibalizumab-Based Bispecific Antibodies.J Acquir Immune Defic Syndr. 2016 Dec 1;73(4):365-373. doi: 10.1097/QAI.0000000000001119. J Acquir Immune Defic Syndr. 2016. PMID: 27792681 Free PMC article.
-
Engineering multi-specific antibodies against HIV-1.Retrovirology. 2018 Aug 29;15(1):60. doi: 10.1186/s12977-018-0439-9. Retrovirology. 2018. PMID: 30157871 Free PMC article. Review.
-
Discoveries and developments of CXCR4-targeted HIV-1 entry inhibitors.Exp Biol Med (Maywood). 2020 Mar;245(5):477-485. doi: 10.1177/1535370220901498. Epub 2020 Feb 4. Exp Biol Med (Maywood). 2020. PMID: 32019336 Free PMC article. Review.
-
Highly potent and broadly neutralizing anti-CD4 trimeric nanobodies inhibit HIV-1 infection by inducing CD4 conformational alteration.Nat Commun. 2024 Aug 13;15(1):6961. doi: 10.1038/s41467-024-51414-6. Nat Commun. 2024. PMID: 39138183 Free PMC article.
-
SARS-CoV-2 resistance to monoclonal antibodies and small-molecule drugs.Cell Chem Biol. 2024 Apr 18;31(4):632-657. doi: 10.1016/j.chembiol.2024.03.008. Cell Chem Biol. 2024. PMID: 38640902 Review.
References
-
- Ashish, Juncadella IJ, Garg R, Boone CD, Anguita J, Krueger JK. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J Biol Chem. 2008;283:2761–2772. - PubMed
-
- Boon L, Holland B, Gordon W, Liu P, Shiau F, Shanahan W, Reimann KA, Fung M. Development of anti-CD4 MAb hu5A8 for treatment of HIV-1 infection: preclinical assessment in non-human primates. Toxicology. 2002;172:191–203. - PubMed
-
- Burkly LC, Olson D, Shapiro R, Winkler G, Rosa JJ, Thomas DW, Williams C, Chisholm P. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal antibody. Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. J Immunol. 1992;149:1779–1787. - PubMed
-
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

