The evolution and diversity of DNA transposons in the genome of the Lizard Anolis carolinensis
- PMID: 21127169
- PMCID: PMC3014272
- DOI: 10.1093/gbe/evq080
The evolution and diversity of DNA transposons in the genome of the Lizard Anolis carolinensis
Abstract
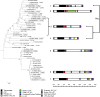
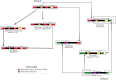
DNA transposons have considerably affected the size and structure of eukaryotic genomes and have been an important source of evolutionary novelties. In vertebrates, DNA transposons are discontinuously distributed due to the frequent extinction and recolonization of these genomes by active elements. We performed a detailed analysis of the DNA transposons in the genome of the lizard Anolis carolinensis, the first non-avian reptile to have its genome sequenced. Elements belonging to six of the previously recognized superfamilies of elements (hAT, Tc1/Mariner, Helitron, PIF/Harbinger, Polinton/Maverick, and Chapaev) were identified. However, only four (hAT, Tc1/Mariner, Helitron, and Chapaev) of these superfamilies have successfully amplified in the anole genome, producing 67 distinct families. The majority (57/67) are nonautonomous and demonstrate an extraordinary diversity of structure, resulting from frequent interelement recombination and incorporation of extraneous DNA sequences. The age distribution of transposon families differs among superfamilies and reveals different dynamics of amplification. Chapaev is the only superfamily to be extinct and is represented only by old copies. The hAT, Tc1/Mariner, and Helitron superfamilies show different pattern of amplification, yet they are predominantly represented by young families, whereas divergent families are exceedingly rare. Although it is likely that some elements, in particular long ones, are subjected to purifying selection and do not reach fixation, the majority of families are neutral and accumulate in the anole genome in large numbers. We propose that the scarcity of old copies in the anole genome results from the rapid decay of elements, caused by a high rate of DNA loss.
Figures




Similar articles
-
Genome-wide analysis of transposable elements in the coffee berry borer Hypothenemus hampei (Coleoptera: Curculionidae): description of novel families.Mol Genet Genomics. 2017 Jun;292(3):565-583. doi: 10.1007/s00438-017-1291-7. Epub 2017 Feb 15. Mol Genet Genomics. 2017. PMID: 28204924
-
The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals.Mol Biol Evol. 2009 Aug;26(8):1811-22. doi: 10.1093/molbev/msp090. Epub 2009 May 6. Mol Biol Evol. 2009. PMID: 19420048
-
Divergent evolution profiles of DD37D and DD39D families of Tc1/mariner transposons in eukaryotes.Mol Phylogenet Evol. 2021 Aug;161:107143. doi: 10.1016/j.ympev.2021.107143. Epub 2021 Mar 10. Mol Phylogenet Evol. 2021. PMID: 33713798
-
Research progress of Tc1/Mariner superfamily.Yi Chuan. 2017 Jan 20;39(1):1-13. doi: 10.16288/j.yczz.16-160. Yi Chuan. 2017. PMID: 28115300 Review.
-
DNA transposons in vertebrate functional genomics.Cell Mol Life Sci. 2005 Mar;62(6):629-41. doi: 10.1007/s00018-004-4232-7. Cell Mol Life Sci. 2005. PMID: 15770416 Review.
Cited by
-
Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution.Genes (Basel). 2020 Jul 21;11(7):827. doi: 10.3390/genes11070827. Genes (Basel). 2020. PMID: 32708239 Free PMC article. Review.
-
Reverse transcriptase and intron number evolution.Stem Cell Investig. 2014 Sep 28;1:17. doi: 10.3978/j.issn.2306-9759.2014.08.01. eCollection 2014. Stem Cell Investig. 2014. PMID: 27358863 Free PMC article.
-
Transposon fingerprinting using low coverage whole genome shotgun sequencing in cacao (Theobroma cacao L.) and related species.BMC Genomics. 2013 Jul 24;14:502. doi: 10.1186/1471-2164-14-502. BMC Genomics. 2013. PMID: 23883295 Free PMC article.
-
HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes.Proc Natl Acad Sci U S A. 2014 Jul 15;111(28):10263-8. doi: 10.1073/pnas.1410068111. Epub 2014 Jun 30. Proc Natl Acad Sci U S A. 2014. PMID: 24982153 Free PMC article.
-
DNA Polymerase Diversity Reveals Multiple Incursions of Polintons During Nematode Evolution.Mol Biol Evol. 2023 Dec 1;40(12):msad274. doi: 10.1093/molbev/msad274. Mol Biol Evol. 2023. PMID: 38069639 Free PMC article.
References
-
- Arkhipova IR. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst Biol. 2006;55:875–885. - PubMed
-
- Craig NL, Craigie R, Gellert M, Lambowitz LM. Mobile DNA II. Washington (DC): American Society for Microbiology Press; 2002.
-
- Duvernell DD, Pryor SR, Adams SM. Teleost fish genomes contain a diverse array of L1 retrotransposon lineages that exhibit a low copy number and high rate of turnover. J Mol Evol. 2004;59:298–308. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

