Evolutionary relationships of wild hominids recapitulated by gut microbial communities
- PMID: 21103409
- PMCID: PMC2982803
- DOI: 10.1371/journal.pbio.1000546
Evolutionary relationships of wild hominids recapitulated by gut microbial communities
Abstract
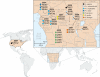
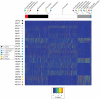
Multiple factors over the lifetime of an individual, including diet, geography, and physiologic state, will influence the microbial communities within the primate gut. To determine the source of variation in the composition of the microbiota within and among species, we investigated the distal gut microbial communities harbored by great apes, as present in fecal samples recovered within their native ranges. We found that the branching order of host-species phylogenies based on the composition of these microbial communities is completely congruent with the known relationships of the hosts. Although the gut is initially and continuously seeded by bacteria that are acquired from external sources, we establish that over evolutionary timescales, the composition of the gut microbiota among great ape species is phylogenetically conserved and has diverged in a manner consistent with vertical inheritance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
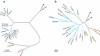
Comment in
-
You aren't always what you eat.PLoS Biol. 2010 Nov 16;8(11):e1001000. doi: 10.1371/journal.pbio.1001000. PLoS Biol. 2010. PMID: 21103411 Free PMC article. No abstract available.
Similar articles
-
Rapid changes in the gut microbiome during human evolution.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16431-5. doi: 10.1073/pnas.1419136111. Epub 2014 Nov 3. Proc Natl Acad Sci U S A. 2014. PMID: 25368157 Free PMC article.
-
Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park.Proc Natl Acad Sci U S A. 2012 Aug 7;109(32):13034-9. doi: 10.1073/pnas.1110994109. Epub 2012 Jul 23. Proc Natl Acad Sci U S A. 2012. PMID: 22826227 Free PMC article.
-
Primate phageomes are structured by superhost phylogeny and environment.Proc Natl Acad Sci U S A. 2021 Apr 13;118(15):e2013535118. doi: 10.1073/pnas.2013535118. Proc Natl Acad Sci U S A. 2021. PMID: 33876746 Free PMC article.
-
How can non-human primates inform evolutionary perspectives on female-biased kinship in humans?Philos Trans R Soc Lond B Biol Sci. 2019 Sep 2;374(1780):20180074. doi: 10.1098/rstb.2018.0074. Epub 2019 Jul 15. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31303156 Free PMC article. Review.
-
Molecular evidence on primate phylogeny from DNA sequences.Am J Phys Anthropol. 1994 May;94(1):3-24. doi: 10.1002/ajpa.1330940103. Am J Phys Anthropol. 1994. PMID: 8042704 Review.
Cited by
-
Captivity humanizes the primate microbiome.Proc Natl Acad Sci U S A. 2016 Sep 13;113(37):10376-81. doi: 10.1073/pnas.1521835113. Epub 2016 Aug 29. Proc Natl Acad Sci U S A. 2016. PMID: 27573830 Free PMC article.
-
Dissecting host-associated communities with DNA barcodes.Philos Trans R Soc Lond B Biol Sci. 2016 Sep 5;371(1702):20150328. doi: 10.1098/rstb.2015.0328. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27481780 Free PMC article. Review.
-
Insights from characterizing extinct human gut microbiomes.PLoS One. 2012;7(12):e51146. doi: 10.1371/journal.pone.0051146. Epub 2012 Dec 12. PLoS One. 2012. PMID: 23251439 Free PMC article.
-
Patterns of gut bacterial colonization in three primate species.PLoS One. 2015 May 13;10(5):e0124618. doi: 10.1371/journal.pone.0124618. eCollection 2015. PLoS One. 2015. PMID: 25970595 Free PMC article.
-
Subsistence strategies in traditional societies distinguish gut microbiomes.Nat Commun. 2015 Mar 25;6:6505. doi: 10.1038/ncomms7505. Nat Commun. 2015. PMID: 25807110 Free PMC article.
References
-
- Inoue R, Ushida K. Vertical and horizontal transmission of intestinal commensal bacteria in the rat model. FEMS Microbiol Ecol. 2003;46:213–219. - PubMed
-
- Favier C. F, De Vos W. M, Akkermans A. D. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. - PubMed
-
- Ley R. E, Peterson D. A, Gordon J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 AI027767/AI/NIAID NIH HHS/United States
- AI065371/AI/NIAID NIH HHS/United States
- R01 GM056120/GM/NIGMS NIH HHS/United States
- GM56120/GM/NIGMS NIH HHS/United States
- R01 AI050529/AI/NIAID NIH HHS/United States
- R01 AI058715/AI/NIAID NIH HHS/United States
- GM74735/GM/NIGMS NIH HHS/United States
- AI27767/AI/NIAID NIH HHS/United States
- AI58715/AI/NIAID NIH HHS/United States
- R21 AI065371/AI/NIAID NIH HHS/United States
- R37 AI050529/AI/NIAID NIH HHS/United States
- R15 GM074735/GM/NIGMS NIH HHS/United States
- AI50529/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

