ModBase, a database of annotated comparative protein structure models, and associated resources
- PMID: 21097780
- PMCID: PMC3013688
- DOI: 10.1093/nar/gkq1091
ModBase, a database of annotated comparative protein structure models, and associated resources
Abstract
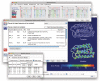
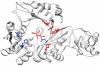
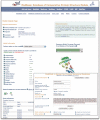
ModBase (http://salilab.org/modbase) is a database of annotated comparative protein structure models. The models are calculated by ModPipe, an automated modeling pipeline that relies primarily on Modeller for fold assignment, sequence-structure alignment, model building and model assessment (http://salilab.org/modeller/). ModBase currently contains 10,355,444 reliable models for domains in 2,421,920 unique protein sequences. ModBase allows users to update comparative models on demand, and request modeling of additional sequences through an interface to the ModWeb modeling server (http://salilab.org/modweb). ModBase models are available through the ModBase interface as well as the Protein Model Portal (http://www.proteinmodelportal.org/). Recently developed associated resources include the SALIGN server for multiple sequence and structure alignment (http://salilab.org/salign), the ModEval server for predicting the accuracy of protein structure models (http://salilab.org/modeval), the PCSS server for predicting which peptides bind to a given protein (http://salilab.org/pcss) and the FoXS server for calculating and fitting Small Angle X-ray Scattering profiles (http://salilab.org/foxs).
Figures
Similar articles
-
ModBase, a database of annotated comparative protein structure models and associated resources.Nucleic Acids Res. 2014 Jan;42(Database issue):D336-46. doi: 10.1093/nar/gkt1144. Epub 2013 Nov 23. Nucleic Acids Res. 2014. PMID: 24271400 Free PMC article.
-
MODBASE: a database of annotated comparative protein structure models and associated resources.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D291-5. doi: 10.1093/nar/gkj059. Nucleic Acids Res. 2006. PMID: 16381869 Free PMC article.
-
MODBASE, a database of annotated comparative protein structure models, and associated resources.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D217-22. doi: 10.1093/nar/gkh095. Nucleic Acids Res. 2004. PMID: 14681398 Free PMC article.
-
MODBASE, a database of annotated comparative protein structure models and associated resources.Nucleic Acids Res. 2009 Jan;37(Database issue):D347-54. doi: 10.1093/nar/gkn791. Epub 2008 Oct 23. Nucleic Acids Res. 2009. PMID: 18948282 Free PMC article.
-
MODBASE, a database of annotated comparative protein structure models.Nucleic Acids Res. 2002 Jan 1;30(1):255-9. doi: 10.1093/nar/30.1.255. Nucleic Acids Res. 2002. PMID: 11752309 Free PMC article.
Cited by
-
Impaired transforming growth factor-β (TGF-β) transcriptional activity and cell proliferation control of a menin in-frame deletion mutant associated with multiple endocrine neoplasia type 1 (MEN1).J Biol Chem. 2012 Mar 9;287(11):8584-97. doi: 10.1074/jbc.M112.341958. Epub 2012 Jan 24. J Biol Chem. 2012. PMID: 22275377 Free PMC article. Clinical Trial.
-
MmTX1 and MmTX2 from coral snake venom potently modulate GABAA receptor activity.Proc Natl Acad Sci U S A. 2015 Feb 24;112(8):E891-900. doi: 10.1073/pnas.1415488112. Epub 2015 Feb 9. Proc Natl Acad Sci U S A. 2015. PMID: 25675485 Free PMC article.
-
Amending HIV Drugs: A Novel Small-Molecule Approach To Target Lupus Anti-DNA Antibodies.J Med Chem. 2016 Oct 13;59(19):8859-8867. doi: 10.1021/acs.jmedchem.6b00694. Epub 2016 Sep 20. J Med Chem. 2016. PMID: 27603688 Free PMC article.
-
New insights into the phylogeny and molecular classification of nicotinamide mononucleotide deamidases.PLoS One. 2013 Dec 5;8(12):e82705. doi: 10.1371/journal.pone.0082705. eCollection 2013. PLoS One. 2013. PMID: 24340054 Free PMC article.
-
Investigation of Antidepressant Properties of Yohimbine by Employing Structure-Based Computational Assessments.Curr Issues Mol Biol. 2021 Oct 27;43(3):1805-1827. doi: 10.3390/cimb43030127. Curr Issues Mol Biol. 2021. PMID: 34889886 Free PMC article.
References
-
- Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. - PubMed
-
- Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

