Phospho.ELM: a database of phosphorylation sites--update 2011
- PMID: 21062810
- PMCID: PMC3013696
- DOI: 10.1093/nar/gkq1104
Phospho.ELM: a database of phosphorylation sites--update 2011
Abstract
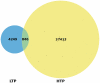
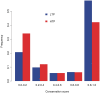
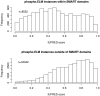
The Phospho.ELM resource (http://phospho.elm.eu.org) is a relational database designed to store in vivo and in vitro phosphorylation data extracted from the scientific literature and phosphoproteomic analyses. The resource has been actively developed for more than 7 years and currently comprises 42,574 serine, threonine and tyrosine non-redundant phosphorylation sites. Several new features have been implemented, such as structural disorder/order and accessibility information and a conservation score. Additionally, the conservation of the phosphosites can now be visualized directly on the multiple sequence alignment used for the score calculation. Finally, special emphasis has been put on linking to external resources such as interaction networks and other databases.
Figures

Similar articles
-
Phospho.ELM: a database of phosphorylation sites--update 2008.Nucleic Acids Res. 2008 Jan;36(Database issue):D240-4. doi: 10.1093/nar/gkm772. Epub 2007 Oct 25. Nucleic Acids Res. 2008. PMID: 17962309 Free PMC article.
-
Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins.BMC Bioinformatics. 2004 Jun 22;5:79. doi: 10.1186/1471-2105-5-79. BMC Bioinformatics. 2004. PMID: 15212693 Free PMC article.
-
Boosting phosphorylation site prediction with sequence feature-based machine learning.Proteins. 2020 Feb;88(2):284-291. doi: 10.1002/prot.25801. Epub 2019 Aug 22. Proteins. 2020. PMID: 31412138
-
From Phosphosites to Kinases.Methods Mol Biol. 2016;1355:307-21. doi: 10.1007/978-1-4939-3049-4_21. Methods Mol Biol. 2016. PMID: 26584935 Review.
-
Serine/threonine/tyrosine phosphorylation regulates DNA binding of bacterial transcriptional regulators.Microbiology (Reading). 2015 Sep;161(9):1720-1729. doi: 10.1099/mic.0.000148. Epub 2015 Jul 23. Microbiology (Reading). 2015. PMID: 26220449 Review.
Cited by
-
Evolutionary Divergence of Phosphorylation to Regulate Interactive Protein Networks in Lower and Higher Species.Int J Mol Sci. 2022 Nov 20;23(22):14429. doi: 10.3390/ijms232214429. Int J Mol Sci. 2022. PMID: 36430905 Free PMC article.
-
From Identification to Characterization of the Multiple Sclerosis Susceptibility Gene CLEC16A.Int J Mol Sci. 2013 Feb 25;14(3):4476-97. doi: 10.3390/ijms14034476. Int J Mol Sci. 2013. PMID: 23439554 Free PMC article.
-
Interpretable deep learning translation of GWAS and multi-omics findings to identify pathobiology and drug repurposing in Alzheimer's disease.Cell Rep. 2022 Nov 29;41(9):111717. doi: 10.1016/j.celrep.2022.111717. Cell Rep. 2022. PMID: 36450252 Free PMC article.
-
Mutational properties of amino acid residues: implications for evolvability of phosphorylatable residues.Philos Trans R Soc Lond B Biol Sci. 2012 Sep 19;367(1602):2584-93. doi: 10.1098/rstb.2012.0076. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22889909 Free PMC article.
-
Protein abundance is key to distinguish promiscuous from functional phosphorylation based on evolutionary information.Philos Trans R Soc Lond B Biol Sci. 2012 Sep 19;367(1602):2594-606. doi: 10.1098/rstb.2012.0078. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22889910 Free PMC article.
References
-
- Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR. Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J. Proteome Res. 2008;7:1346–1351. - PubMed
-
- Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. - PubMed
-
- Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. - PubMed
-
- Lemeer S, Heck AJ. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 2009;13:414–420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

