Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation
- PMID: 21060799
- PMCID: PMC2966431
- DOI: 10.1371/journal.pone.0013769
Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation
Abstract
Background: Methylation of residues in histone tails is part of a network that regulates gene expression. JmjC domain containing proteins catalyze the oxidative removal of methyl groups on histone lysine residues. Here, we report studies to test the involvement of Jumonji domain-containing protein 6 (Jmjd6) in histone lysine demethylation. Jmjd6 has recently been shown to hydroxylate RNA splicing factors and is known to be essential for the differentiation of multiple tissues and cells during embryogenesis. However, there have been conflicting reports as to whether Jmjd6 is a histone-modifying enzyme.
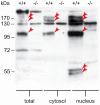
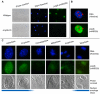
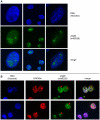
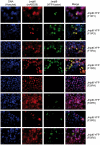
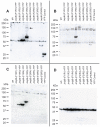
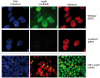
Methodology/principal findings: Immunolocalization studies reveal that Jmjd6 is distributed throughout the nucleoplasm outside of regions containing heterochromatic DNA, with occasional localization in nucleoli. During mitosis, Jmjd6 is excluded from the nucleus and reappears in the telophase of the cell cycle. Western blot analyses confirmed that Jmjd6 forms homo-multimers of different molecular weights in the nucleus and cytoplasm. A comparison of mono-, di-, and tri-methylation states of H3K4, H3K9, H3K27, H3K36, and H4K20 histone residues in wildtype and Jmjd6-knockout cells indicate that Jmjd6 is not involved in the demethylation of these histone lysine residues. This is further supported by overexpression of enzymatically active and inactive forms of Jmjd6 and subsequent analysis of histone methylation patterns by immunocytochemistry and western blot analysis. Finally, treatment of cells with RNase A and DNase I indicate that Jmjd6 may preferentially associate with RNA/RNA complexes and less likely with chromatin.
Conclusions/significance: Taken together, our results provide further evidence that Jmjd6 is unlikely to be involved in histone lysine demethylation. We confirmed that Jmjd6 forms multimers and showed that nuclear localization of the protein involves association with a nucleic acid matrix.
Conflict of interest statement
Figures
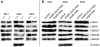
Similar articles
-
Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6).J Biol Chem. 2013 Mar 1;288(9):6053-62. doi: 10.1074/jbc.M112.433284. Epub 2013 Jan 9. J Biol Chem. 2013. PMID: 23303181 Free PMC article.
-
The hydroxylation activity of Jmjd6 is required for its homo-oligomerization.J Cell Biochem. 2012 May;113(5):1663-70. doi: 10.1002/jcb.24035. J Cell Biochem. 2012. PMID: 22189873
-
Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: role of Jumonji C-domain containing protein 6 in RHA demethylation.Virology. 2014 Mar;452-453:1-11. doi: 10.1016/j.virol.2013.12.040. Epub 2014 Jan 23. Virology. 2014. PMID: 24606677
-
Insights into Jumonji C-domain containing protein 6 (JMJD6): a multifactorial role in foot-and-mouth disease virus replication in cells.Virus Genes. 2017 Jun;53(3):340-351. doi: 10.1007/s11262-017-1449-8. Epub 2017 Mar 31. Virus Genes. 2017. PMID: 28364140 Review.
-
The oxygenase Jmjd6--a case study in conflicting assignments.Biochem J. 2015 Jun 1;468(2):191-202. doi: 10.1042/BJ20150278. Biochem J. 2015. PMID: 25997831 Review.
Cited by
-
Protein arginine methylation/demethylation and cancer.Oncotarget. 2016 Oct 11;7(41):67532-67550. doi: 10.18632/oncotarget.11376. Oncotarget. 2016. PMID: 27556302 Free PMC article. Review.
-
Dynamic Arginine Methylation of Tumor Necrosis Factor (TNF) Receptor-associated Factor 6 Regulates Toll-like Receptor Signaling.J Biol Chem. 2015 Sep 4;290(36):22236-49. doi: 10.1074/jbc.M115.653543. Epub 2015 Jul 28. J Biol Chem. 2015. PMID: 26221041 Free PMC article.
-
Promotion of adipogenesis by JMJD6 requires the AT hook-like domain and is independent of its catalytic function.PLoS One. 2019 Aug 20;14(8):e0216015. doi: 10.1371/journal.pone.0216015. eCollection 2019. PLoS One. 2019. PMID: 31430278 Free PMC article.
-
Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1.J Biol Chem. 2017 Nov 17;292(46):18886-18896. doi: 10.1074/jbc.M117.800706. Epub 2017 Sep 27. J Biol Chem. 2017. PMID: 28972166 Free PMC article.
-
Protein arginine methyltransferases and cancer.Nat Rev Cancer. 2013 Jan;13(1):37-50. doi: 10.1038/nrc3409. Epub 2012 Dec 13. Nat Rev Cancer. 2013. PMID: 23235912 Review.
References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

