Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase
- PMID: 21059884
- PMCID: PMC3018298
- DOI: 10.1534/genetics.110.124347
Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase
Abstract
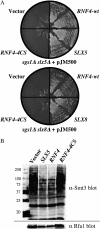
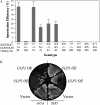
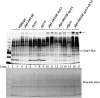
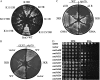
Saccharomyces cerevisiae cells lacking the Slx5-Slx8 SUMO-targeted Ub ligase display increased levels of sumoylated and polysumoylated proteins, and they are inviable in the absence of the Sgs1 DNA helicase. One explanation for this inviability is that one or more sumoylated proteins accumulate to toxic levels in sgs1Δ slx5Δ cells. To address this possibility, we isolated a second-site suppressor of sgs1Δ slx5Δ synthetic lethality and identified it as an allele of the ULP2 SUMO isopeptidase. The suppressor, ulp2-D623H, behaved like the ulp2Δ allele in its sensitivity to heat, DNA replication stress, and DNA damage. Surprisingly, deletion of ULP2, which is known to promote the accumulation of poly-SUMO chains, suppressed sgs1Δ slx5Δ synthetic lethality and the slx5Δ sporulation defect. Further, ulp2Δ's growth sensitivities were found to be suppressed in ulp2Δ slx5Δ double mutants. This mutual suppression indicates that SLX5-SLX8 and ULP2 interact antagonistically. However, the suppressed strain sgs1Δ slx5Δ ulp2-D623H displayed even higher levels of sumoylated proteins than the corresponding double mutants. Thus, sgs1Δ slx5Δ synthetic lethality cannot be due simply to high levels of bulk sumoylated proteins. We speculate that the loss of ULP2 suppresses the toxicity of the sumoylated proteins that accumulate in slx5Δ-slx8Δ cells by permitting the extension of poly-SUMO chains on specific target proteins. This additional modification might attenuate the activity of the target proteins or channel them into alternative pathways for proteolytic degradation. In support of this latter possibility we find that the WSS1 isopeptidase is required for suppression by ulp2Δ.
Figures






Similar articles
-
SUMO Pathway Modulation of Regulatory Protein Binding at the Ribosomal DNA Locus in Saccharomyces cerevisiae.Genetics. 2016 Apr;202(4):1377-94. doi: 10.1534/genetics.116.187252. Epub 2016 Feb 2. Genetics. 2016. PMID: 26837752 Free PMC article.
-
A Lysine Desert Protects a Novel Domain in the Slx5-Slx8 SUMO Targeted Ub Ligase To Maintain Sumoylation Levels in Saccharomyces cerevisiae.Genetics. 2017 Aug;206(4):1807-1821. doi: 10.1534/genetics.117.202697. Epub 2017 May 26. Genetics. 2017. PMID: 28550017 Free PMC article.
-
Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase.Mol Cell Biol. 2010 Aug;30(15):3737-48. doi: 10.1128/MCB.01649-09. Epub 2010 Jun 1. Mol Cell Biol. 2010. PMID: 20516210 Free PMC article.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
-
Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases.Nucleic Acids Res. 2017 Mar 17;45(5):2242-2261. doi: 10.1093/nar/gkw1369. Nucleic Acids Res. 2017. PMID: 28115630 Free PMC article. Review.
Cited by
-
SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability.Int J Mol Sci. 2021 May 20;22(10):5391. doi: 10.3390/ijms22105391. Int J Mol Sci. 2021. PMID: 34065507 Free PMC article. Review.
-
SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):16586-91. doi: 10.1073/pnas.1417548111. Epub 2014 Nov 5. Proc Natl Acad Sci U S A. 2014. PMID: 25378699 Free PMC article.
-
SUMO Pathway Modulation of Regulatory Protein Binding at the Ribosomal DNA Locus in Saccharomyces cerevisiae.Genetics. 2016 Apr;202(4):1377-94. doi: 10.1534/genetics.116.187252. Epub 2016 Feb 2. Genetics. 2016. PMID: 26837752 Free PMC article.
-
STUbLs in chromatin and genome stability.Biopolymers. 2013 Feb;99(2):146-54. doi: 10.1002/bip.22125. Biopolymers. 2013. PMID: 23175389 Free PMC article. Review.
-
DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.Mol Cell Biol. 2011 Jun;31(11):2299-310. doi: 10.1128/MCB.05188-11. Epub 2011 Mar 28. Mol Cell Biol. 2011. PMID: 21444718 Free PMC article.
References
-
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner and S. J. Elledge, 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9 1169–1182. - PubMed
-
- Bylebyl, G. R., I. Belichenko and E. S. Johnson, 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278 44113–44120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

