Molecular signals of epigenetic states
- PMID: 21030644
- PMCID: PMC3772643
- DOI: 10.1126/science.1191078
Molecular signals of epigenetic states
Abstract
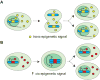
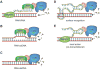
Epigenetic signals are responsible for the establishment, maintenance, and reversal of metastable transcriptional states that are fundamental for the cell's ability to "remember" past events, such as changes in the external environment or developmental cues. Complex epigenetic states are orchestrated by several converging and reinforcing signals, including transcription factors, noncoding RNAs, DNA methylation, and histone modifications. Although all of these pathways modulate transcription from chromatin in vivo, the mechanisms by which epigenetic information is transmitted through cell division remain unclear. Because epigenetic states are metastable and change in response to the appropriate signals, a deeper understanding of their molecular framework will allow us to tackle the dysregulation of epigenetics in disease.
Figures

Similar articles
-
Three-dimensional regulation of transcription.Protein Cell. 2015 Apr;6(4):241-53. doi: 10.1007/s13238-015-0135-7. Epub 2015 Feb 12. Protein Cell. 2015. PMID: 25670626 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Signals and combinatorial functions of histone modifications.Annu Rev Biochem. 2011;80:473-99. doi: 10.1146/annurev-biochem-061809-175347. Annu Rev Biochem. 2011. PMID: 21529160 Review.
-
Decoding liver injury: A regulatory role for histone modifications.Int J Biochem Cell Biol. 2015 Oct;67:188-93. doi: 10.1016/j.biocel.2015.03.009. Epub 2015 Mar 20. Int J Biochem Cell Biol. 2015. PMID: 25801055 Review.
-
Epigenetic control of RNA polymerase I transcription in mammalian cells.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):393-404. doi: 10.1016/j.bbagrm.2012.10.004. Epub 2012 Oct 12. Biochim Biophys Acta. 2013. PMID: 23063748 Review.
Cited by
-
Epigenetic perturbations in aging stem cells.Mamm Genome. 2016 Aug;27(7-8):396-406. doi: 10.1007/s00335-016-9645-8. Epub 2016 May 26. Mamm Genome. 2016. PMID: 27229519 Free PMC article. Review.
-
Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer.Biomolecules. 2016 Apr 28;6(2):25. doi: 10.3390/biom6020025. Biomolecules. 2016. PMID: 27136596 Free PMC article. Review.
-
Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes.Epigenetics. 2012 Sep;7(9):969-75. doi: 10.4161/epi.21337. Epub 2012 Aug 7. Epigenetics. 2012. PMID: 22869105 Free PMC article.
-
HIstome--a relational knowledgebase of human histone proteins and histone modifying enzymes.Nucleic Acids Res. 2012 Jan;40(Database issue):D337-42. doi: 10.1093/nar/gkr1125. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140112 Free PMC article.
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization.Cell Death Differ. 2024 Jan 16. doi: 10.1038/s41418-023-01254-6. Online ahead of print. Cell Death Differ. 2024. PMID: 38228801 Review.
References
-
- Ringrose L, Paro R. Annu Rev Genet. 2004 Dec 1;38:413. - PubMed
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman and Hall/CRC; 2006.
-
- Brennecke J, et al. Cell. 2007 Mar;128:1089. - PubMed
-
- Talbert PB, Henikoff S. Nat Rev Mol Cell Biol. 2010 Apr;11:264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

