Blood peptidome-degradome profile of breast cancer
- PMID: 20976186
- PMCID: PMC2956627
- DOI: 10.1371/journal.pone.0013133
Blood peptidome-degradome profile of breast cancer
Abstract
Background: Cancer invasion and metastasis are closely associated with activities within the degradome; however, little is known about whether these activities can be detected in the blood of cancer patients.
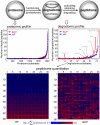
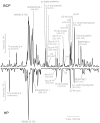
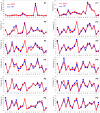
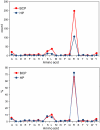
Methodology and principal findings: The peptidome-degradome profiles of pooled blood plasma sampled from 15 breast cancer patients (BCP) and age, race, and menopausal status matched control healthy persons (HP) were globally characterized using advanced comprehensive separations combined with tandem Fourier transform mass spectrometry and new data analysis approaches that facilitated top-down peptidomic analysis. The BCP pool displayed 71 degradome protein substrates that encompassed 839 distinct peptidome peptides. In contrast, the HP 50 degradome substrates found encompassed 425 peptides. We find that the ratios of the peptidome peptide relative abundances can vary as much as >4000 fold between BCP and HP. The experimental results also show differential degradation of substrates in the BCP sample in their functional domains, including the proteolytic and inhibitory sites of the plasmin-antiplasmin and thrombin-antithrombin systems, the main chains of the extracellular matrix protection proteins, the excessive degradation of innate immune system key convertases and membrane attack complex components, as well as several other cancer suppressor proteins.
Conclusions: Degradomics-peptidomics profiling of blood plasma is highly sensitive to changes not evidenced by conventional bottom-up proteomics and potentially provides unique signatures of possible diagnostic utility.
Conflict of interest statement
Figures


Similar articles
-
The Peptidome Comes of Age: Mass Spectrometry-Based Characterization of the Circulating Cancer Peptidome.Enzymes. 2017;42:27-64. doi: 10.1016/bs.enz.2017.08.003. Epub 2017 Oct 10. Enzymes. 2017. PMID: 29054270 Review.
-
Characterization of the Low-Molecular-Weight Human Plasma Peptidome.Methods Mol Biol. 2017;1619:63-79. doi: 10.1007/978-1-4939-7057-5_6. Methods Mol Biol. 2017. PMID: 28674878
-
Improving collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD) fourier transform MS/MS degradome-peptidome identifications using high accuracy mass information.J Proteome Res. 2012 Feb 3;11(2):668-77. doi: 10.1021/pr200597j. Epub 2011 Dec 1. J Proteome Res. 2012. PMID: 22054047 Free PMC article.
-
Strategy for degradomic-peptidomic analysis of human blood plasma.J Proteome Res. 2010 May 7;9(5):2339-46. doi: 10.1021/pr901083m. J Proteome Res. 2010. PMID: 20377236 Free PMC article.
-
The emerging role of the peptidome in biomarker discovery and degradome profiling.Biol Chem. 2015 Mar;396(3):185-92. doi: 10.1515/hsz-2014-0207. Biol Chem. 2015. PMID: 25229414 Review.
Cited by
-
Mass spectrometry-based proteomics: existing capabilities and future directions.Chem Soc Rev. 2012 May 21;41(10):3912-28. doi: 10.1039/c2cs15331a. Epub 2012 Apr 13. Chem Soc Rev. 2012. PMID: 22498958 Free PMC article. Review.
-
Characterization of the Ovarian Tumor Peptidome.Vitam Horm. 2018;107:515-531. doi: 10.1016/bs.vh.2018.01.020. Epub 2018 Feb 22. Vitam Horm. 2018. PMID: 29544642 Free PMC article.
-
Clinical ApoA-IV amyloid is associated with fibrillogenic signal sequence.J Pathol. 2021 Nov;255(3):311-318. doi: 10.1002/path.5770. Epub 2021 Aug 27. J Pathol. 2021. PMID: 34331462 Free PMC article.
-
Carrying yourself: self antigen composition of the lymphatic fluid.Lymphat Res Biol. 2013 Sep;11(3):149-54. doi: 10.1089/lrb.2013.0009. Epub 2013 Sep 11. Lymphat Res Biol. 2013. PMID: 24024574 Free PMC article.
-
Contribution of the plasma and lymph Degradome and Peptidome to the MHC Ligandome.Immunogenetics. 2019 Mar;71(3):203-216. doi: 10.1007/s00251-018-1093-z. Epub 2018 Oct 20. Immunogenetics. 2019. PMID: 30343358 Free PMC article. Review.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brünner N, et al. National academy of clinical biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clin Chem. 2008;54:e11–e79. - PubMed
-
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, et al. American society of clinical oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. - PubMed
-
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

