Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate
- PMID: 20961956
- PMCID: PMC3045611
- DOI: 10.1093/nar/gkq974
Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate
Abstract
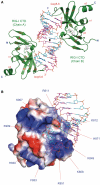
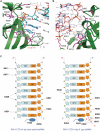
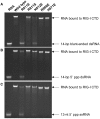
RIG-I recognizes molecular patterns in viral RNA to regulate the induction of type I interferons. The C-terminal domain (CTD) of RIG-I exhibits high affinity for 5' triphosphate (ppp) dsRNA as well as blunt-ended dsRNA. Structures of RIG-I CTD bound to 5'-ppp dsRNA showed that RIG-I recognizes the termini of dsRNA and interacts with the ppp through electrostatic interactions. However, the structural basis for the recognition of non-phosphorylated dsRNA by RIG-I is not fully understood. Here, we show that RIG-I CTD binds blunt-ended dsRNA in a different orientation compared to 5' ppp dsRNA and interacts with both strands of the dsRNA. Overlapping sets of residues are involved in the recognition of blunt-ended dsRNA and 5' ppp dsRNA. Mutations at the RNA-binding surface affect RNA binding and signaling by RIG-I. These results provide the mechanistic basis for how RIG-I recognizes different RNA ligands.
Figures

Similar articles
-
The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain.Structure. 2010 Aug 11;18(8):1032-43. doi: 10.1016/j.str.2010.05.007. Epub 2010 Jul 15. Structure. 2010. PMID: 20637642 Free PMC article.
-
Structural basis of RNA recognition and activation by innate immune receptor RIG-I.Nature. 2011 Sep 25;479(7373):423-7. doi: 10.1038/nature10537. Nature. 2011. PMID: 21947008 Free PMC article.
-
Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5.Arch Biochem Biophys. 2009 Aug 1;488(1):23-33. doi: 10.1016/j.abb.2009.06.008. Epub 2009 Jun 14. Arch Biochem Biophys. 2009. PMID: 19531363
-
A structure-based model of RIG-I activation.RNA. 2012 Dec;18(12):2118-27. doi: 10.1261/rna.035949.112. Epub 2012 Nov 1. RNA. 2012. PMID: 23118418 Free PMC article. Review.
-
Structural mechanism of RNA recognition by the RIG-I-like receptors.Immunity. 2008 Aug 15;29(2):178-81. doi: 10.1016/j.immuni.2008.07.009. Immunity. 2008. PMID: 18701081 Review.
Cited by
-
The battle between host and SARS-CoV-2: Innate immunity and viral evasion strategies.Mol Ther. 2022 May 4;30(5):1869-1884. doi: 10.1016/j.ymthe.2022.02.014. Epub 2022 Feb 14. Mol Ther. 2022. PMID: 35176485 Free PMC article. Review.
-
RIG-I Uses an ATPase-Powered Translocation-Throttling Mechanism for Kinetic Proofreading of RNAs and Oligomerization.Mol Cell. 2018 Oct 18;72(2):355-368.e4. doi: 10.1016/j.molcel.2018.08.021. Epub 2018 Sep 27. Mol Cell. 2018. PMID: 30270105 Free PMC article.
-
High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5.Nucleic Acids Res. 2015 Jan;43(2):1216-30. doi: 10.1093/nar/gku1329. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539915 Free PMC article.
-
Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction.J Virol. 2012 Apr;86(7):3411-21. doi: 10.1128/JVI.06405-11. Epub 2012 Feb 1. J Virol. 2012. PMID: 22301134 Free PMC article.
-
The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-β response.J Biol Chem. 2019 Apr 19;294(16):6430-6438. doi: 10.1074/jbc.RA118.005868. Epub 2019 Feb 25. J Biol Chem. 2019. PMID: 30804210 Free PMC article.
References
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. - PubMed
-
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
-
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

