An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy
- PMID: 20957184
- PMCID: PMC2950124
- DOI: 10.1371/journal.pbio.1000502
An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy
Abstract
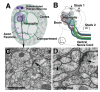
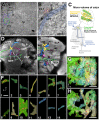
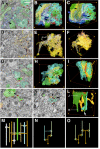
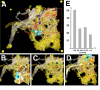
The analysis of microcircuitry (the connectivity at the level of individual neuronal processes and synapses), which is indispensable for our understanding of brain function, is based on serial transmission electron microscopy (TEM) or one of its modern variants. Due to technical limitations, most previous studies that used serial TEM recorded relatively small stacks of individual neurons. As a result, our knowledge of microcircuitry in any nervous system is very limited. We applied the software package TrakEM2 to reconstruct neuronal microcircuitry from TEM sections of a small brain, the early larval brain of Drosophila melanogaster. TrakEM2 enables us to embed the analysis of the TEM image volumes at the microcircuit level into a light microscopically derived neuro-anatomical framework, by registering confocal stacks containing sparsely labeled neural structures with the TEM image volume. We imaged two sets of serial TEM sections of the Drosophila first instar larval brain neuropile and one ventral nerve cord segment, and here report our first results pertaining to Drosophila brain microcircuitry. Terminal neurites fall into a small number of generic classes termed globular, varicose, axiform, and dendritiform. Globular and varicose neurites have large diameter segments that carry almost exclusively presynaptic sites. Dendritiform neurites are thin, highly branched processes that are almost exclusively postsynaptic. Due to the high branching density of dendritiform fibers and the fact that synapses are polyadic, neurites are highly interconnected even within small neuropile volumes. We describe the network motifs most frequently encountered in the Drosophila neuropile. Our study introduces an approach towards a comprehensive anatomical reconstruction of neuronal microcircuitry and delivers microcircuitry comparisons between vertebrate and insect neuropile.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Drosophila brain development: closing the gap between a macroarchitectural and microarchitectural approach.Cold Spring Harb Symp Quant Biol. 2009;74:235-48. doi: 10.1101/sqb.2009.74.037. Epub 2009 Dec 22. Cold Spring Harb Symp Quant Biol. 2009. PMID: 20028843 Free PMC article.
-
As-rigid-as-possible mosaicking and serial section registration of large ssTEM datasets.Bioinformatics. 2010 Jun 15;26(12):i57-63. doi: 10.1093/bioinformatics/btq219. Bioinformatics. 2010. PMID: 20529937 Free PMC article.
-
Different classes of input and output neurons reveal new features in microglomeruli of the adult Drosophila mushroom body calyx.J Comp Neurol. 2012 Jul 1;520(10):2185-201. doi: 10.1002/cne.23037. J Comp Neurol. 2012. PMID: 22237598
-
Beyond counts and shapes: studying pathology of dendritic spines in the context of the surrounding neuropil through serial section electron microscopy.Neuroscience. 2013 Oct 22;251:75-89. doi: 10.1016/j.neuroscience.2012.04.061. Epub 2012 May 1. Neuroscience. 2013. PMID: 22561733 Free PMC article. Review.
-
New developments in electron microscopy for serial image acquisition of neuronal profiles.Microscopy (Oxf). 2015 Feb;64(1):27-36. doi: 10.1093/jmicro/dfu111. Epub 2015 Jan 5. Microscopy (Oxf). 2015. PMID: 25564566 Review.
Cited by
-
Software tool for 3D extraction of germinal centers.BMC Bioinformatics. 2013;14 Suppl 6(Suppl 6):S5. doi: 10.1186/1471-2105-14-S6-S5. Epub 2013 Apr 17. BMC Bioinformatics. 2013. PMID: 23735122 Free PMC article.
-
Assessing CB1 Expression in the Brain by Immunohistochemical Methods: Light, Confocal, and Electron Microscopy.Methods Mol Biol. 2023;2576:407-424. doi: 10.1007/978-1-0716-2728-0_34. Methods Mol Biol. 2023. PMID: 36152206
-
Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae.Integr Comp Biol. 2013 Jul;53(1):7-16. doi: 10.1093/icb/ict046. Epub 2013 May 10. Integr Comp Biol. 2013. PMID: 23667045 Free PMC article.
-
Large-scale automatic reconstruction of neuronal processes from electron microscopy images.Med Image Anal. 2015 May;22(1):77-88. doi: 10.1016/j.media.2015.02.001. Epub 2015 Mar 2. Med Image Anal. 2015. PMID: 25791436 Free PMC article.
-
The Active Segmentation Platform for Microscopic Image Classification and Segmentation.Brain Sci. 2021 Dec 14;11(12):1645. doi: 10.3390/brainsci11121645. Brain Sci. 2021. PMID: 34942947 Free PMC article.
References
-
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Adv Exp Med Biol. 2008;628:1–31. - PubMed
-
- Hartenstein V, Cardona A, Pereanu W, Younossi-Hartenstein A. Modeling the developing Drosophila brain: rationale, technique and application. BioScience. 2008;58:823–836.
-
- Cropley V. L, Fujita M, Innis R. B, Nathan P. J. Molecular imaging of the dopaminergic system and its association with human cognitive function. Biol Psychiatry. 2006;59:898–907. - PubMed
-
- May A. Neuroimaging: visualising the brain in pain. Neurol Sci. 2007;28(Suppl 2):S101–S107. - PubMed
-
- Shibasaki H. Human brain mapping: hemodynamic response and electrophysiology. Clin Neurophysiol. 2008;119:731–743. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

